













| General Information: |
|
| Name(s) found: |
HSFB4_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 348 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
asymmetric cell division
[IMP]
|
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAMMVENSYG GYGGGGGERI QLMVEGQGKA VPAPFLTKTY QLVDDPATDH VVSWGDDDTT 60
61 FVVWRPPEFA RDLLPNYFKH NNFSSFVRQL NTYGFRKIVP DRWEFANEFF KRGEKHLLCE 120
121 IHRRKTSQMI PQQHSPFMSH HHAPPQIPFS GGSFFPLPPP RVTTPEEDHY WCDDSPPSRP 180
181 RVIPQQIDTA AQVTALSEDN ERLRRSNTVL MSELAHMKKL YNDIIYFVQN HVKPVAPSNN 240
241 SSYLSSFLQK QQQQQPPTLD YYNTATVNAT NLNALNSSPP TSQSSITVLE DDHTNHHDQS 300
301 NMRKTKLFGV SLPSSKKRSH HFSDQTSKTR LVLDQSDLAL NLMTASTR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
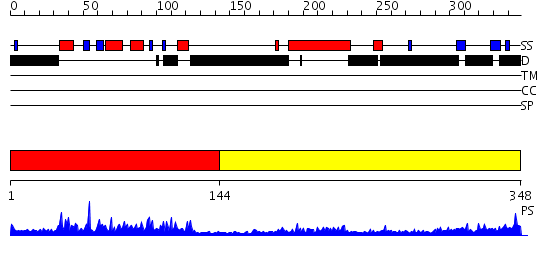
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] | 33.0 | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | |
| 2 | View Details | [144..348] | 3.212996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)