













| General Information: |
|
| Name(s) found: |
DRL17_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 762 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endomembrane system
[IEA]
|
| Biological Process: |
apoptosis
[IEA]
defense response [ISS] |
| Molecular Function: |
ATP binding
[IEA]
protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGCCFSVQFS FDDQTLVRIF NFLCGNINRN SFGVEERPTQ PTIGQEEMLE KAWNRLMEDR 60
61 VGIMGLHGMG GVGKTTLFKK IHNKFAKMSS RFDIVIWIVV SKGAKLSKLQ EDIAEKLHLC 120
121 DDLWKNKNES DKATDIHRVL KGKRFVLMLD DIWEKVDLEA IGVPYPSEVN KCKVAFTTRD 180
181 QKVCGEMGDH KPMQVKCLEP EDAWELFKNK VGDNTLRSDP VIVELAREVA QKCRGLPLAL 240
241 SVIGETMASK TMVQEWEHAI DVLTRSAAEF SNMGNKILPI LKYSYDSLGD EHIKSCFLYC 300
301 ALFPEDDEIY NEKLIDYWIC EGFIGEDQVI KRARNKGYEM LGTLTLANLL TKVGTEHVVM 360
361 HDVVREMALW IASDFGKQKE NFVVRARVGL HERPEAKDWG AVRRMSLMDN HIEEITCESK 420
421 CSELTTLFLQ SNQLKNLSGE FIRYMQKLVV LDLSYNRDFN KLPEQISGLV SLQFLDLSNT 480
481 SIKQLPVGLK KLKKLTFLNL AYTVRLCSIS GISRLLSLRL LRLLGSKVHG DASVLKELQK 540
541 LQNLQHLAIT LSAELSLNQR LANLISILGI EGFLQKPFDL SFLASMENLS SLWVKNSYFS 600
601 EIKCRESETA SSYLRINPKI PCFTNLSRLG LSKCHSIKDL TWILFAPNLV YLYIEDSREV 660
661 GEIINKEKAT NLTSITPFLK LERLILYNLP KLESIYWSPL HFPRLLIIHV LDCPKLRKLP 720
721 LNATSVPLVE EFQIRMYPPG LGNELEWEDE DTKNRFVLSI KK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
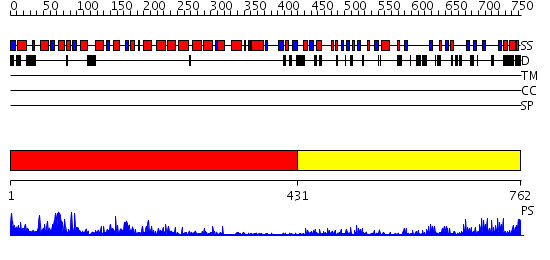
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..430] | 15.09691 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 2 | View Details | [431..762] | 10.0 | No description for 2z63A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)