













| General Information: |
|
| Name(s) found: |
PUB58_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 437 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ubiquitin ligase complex
[IEA]
|
| Biological Process: |
RNA metabolic process
[IEA]
protein ubiquitination [IEA] |
| Molecular Function: |
ubiquitin-protein ligase activity
[IEA]
protein binding [IEA] binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVENSYVLFA RLCVELFHEL PPLPSDESHG DITTFERLFL RKCRIELENA SPKTEHLPLV 60
61 YVDETNYYRF IKTVRVLAEV YKNSKITETT RKSMIQVLMN PILPPERITD AMNLFRSIIG 120
121 KLADFHFSDE KFNQLVRSSR VVELEGNYNE EVKLRKEAED ALAMKKEDVE MMEQLLESYK 180
181 EEQGKLQLQA KALEHKLEAE LRHRKETETL LAIERDRIEK VKIQLETVEN EIDNTRLKAE 240
241 EFERKYEGEM ILRRESEIAL EKEKKELEEV KLKLETYERE QENLSSEVRT WQDKYEQESS 300
301 LRKLSEYALS REQEELQIVK GLLEFYNGEA DAMREERDKA LKTAKEQMEK RQPPSSFFCP 360
361 ITQASPNLYK KMITCITLNL EVMKDPHFAA DGFTYEAESI RKWLSTGHQT SPMTNLRLSH 420
421 LTLVPNRALR SAIEELV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
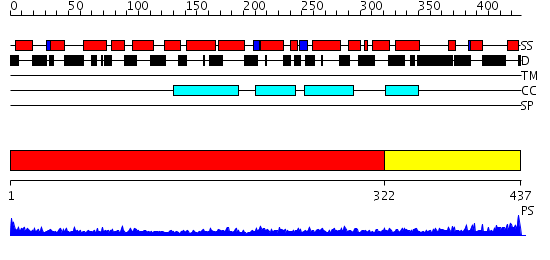
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..321] | 25.69897 | Heavy meromyosin subfragment | |
| 2 | View Details | [322..437] | 4.0 | Crystal structure of the CHIP U-box E3 ubiquitin ligase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)