













| General Information: |
|
| Name(s) found: |
BSL2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1018 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
protein serine/threonine phosphatase activity
[ISS]
phosphoprotein phosphatase activity [IEA] iron ion binding [IEA] manganese ion binding [IEA] hydrolase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDEDSSMVAD NDQDREFQSL DGGQSPSPME RETPQQMNDQ SPPPEGGSVP TPPPSDPNPA 60
61 TSQQQAAAVV GQEQQPALVV GPRCAPTYSV VDAMMDKKED GPGPRCGHTL TAVPAVGDEG 120
121 TPGYIGPRLV LFGGATALEG NSGGTGTPTS AGSAGIRLAG ATADVHCYDV LSNKWTRLTP 180
181 FGEPPTPRAA HVATAVGTMV VIQGGIGPAG LSAEDLHVLD LTQQRPRWHR VVVQGPGPGP 240
241 RYGHVMALVG QRYLMAIGGN DGKRPLADVW ALDTAAKPYE WRKLEPEGEG PPPCMYATAS 300
301 ARSDGLLLLC GGRDANSVPL ASAYGLAKHR DGRWEWAIAP GVSPSSRYQH AAVFVNARLH 360
361 VSGGALGGGR MVEDSSSVAV LDTAAGVWCD TKSVVTSPRT GRYSADAAGG DASVELTRRC 420
421 RHAAAAVGDL IFIYGGLRGG VLLDDLLVAE DLAAAETTYA ASHAAAAAAT NSPPGRLPGR 480
481 YGFSDERNRE LSESAADGAV VLGSPVAPPV NGDMHTDISP ENALLPGTRR TNKGVEYLVE 540
541 ASAAEAEAIS ATLAAAKARQ VNGEVELPDR DCGAEATPSG KPTFSLIKPD SMGSMSVTPA 600
601 GIRLHHRAVV VAAETGGALG GMVRQLSIDQ FENEGRRVSY GTPESATAAR KLLDRQMSIN 660
661 SVPKKVIAHL LKPRGWKPPV RRQFFLDCNE IADLCDSAER IFASEPTVLQ LKAPIKIFGD 720
721 LHGQFGDLMR LFDEYGSPST AGDISYIDYL FLGDYVDRGQ HSLETISLLL ALKVEYQHNV 780
781 HLIRGNHEAA DINALFGFRI ECIERMGERD GIWVWHRINR LFNWLPLAAS IEKKIICMHG 840
841 GIGRSINHVE QIENIQRPIT MEAGSIVLMD LLWSDPTEND SVEGLRPNAR GPGLVTFGPD 900
901 RVMEFCNNND LQLIVRAHEC VMDGFERFAQ GHLITLFSAT NYCGTANNAG AILVLGRDLV 960
961 VVPKLIHPLP PALSSPETSP ERHIEDTWMQ ELNANRPATP TRGRPQNSND RGGSLAWM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
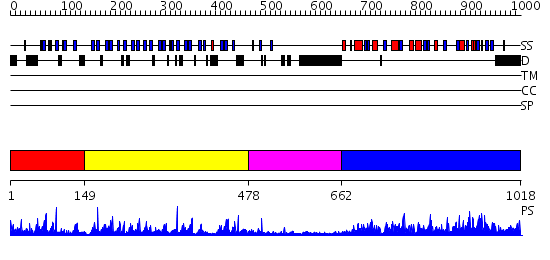
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..148] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [149..477] | 50.522879 | 1.35 angstrom structure of the Kelch domain of Keap1 | |
| 3 | View Details | [478..661] | 41.045757 | Structural basis for the defects of human lung cancer somatic mutations in the repression activity of Keap1 on Nrf2 | |
| 4 | View Details | [662..1018] | 133.0 | Complex between protein ser/thr phosphatase-1 (delta) and the myosin phosphatase targeting subunit 1 (MYPT1) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)