













| General Information: |
|
| Name(s) found: |
SHGR2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 933 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plant-type vacuole membrane
[IDA]
vacuole [IDA] |
| Biological Process: |
negative gravitropism
[IMP]
detection of gravity [IMP] amyloplast organization [IMP] gravitropism [IMP] |
| Molecular Function: |
phospholipase A1 activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEDRETHLGT REVNETSPDL LKNTPSNIAR LEDVIEQCHG RQKYLAQTRS PSDGSDVRWY 60
61 FCKVPLAENE LAASVPRTDV VGKSEYFRFG MRDSLAIEAS FLQREDELLS LWWKEYAECS 120
121 EGPKLQVNSK KKSIETPSEA SVSSSLYEVE EERVGVPVKG GLYEVDLVRR HCFPVYWNGD 180
181 NRRVLRGHWF ARKGGLDWLP IPETVSEQLE VAYRNKVWRR RSFQPSGLFA ARIDLQGSSL 240
241 GLHALFTGED DTWEAWLNVD PSGFSGIVGY TGNGIKLRRG YAGSYSPKPT QEELRQQKEE 300
301 EMDDYCSQVP VRHLVFMVHG IGQKGEKSNL VDDVGNFRQI TAALAERHLT SHQLSTQRVL 360
361 FIPCQWRKGL KLSGEAAVDK CTLDGVRRFR EMLSATVHDV LYYMSPIYCQ AIIDSVSKQL 420
421 NRLYLKFLKR NPDYVGKISI YGHSLGSVLS YDILCHQHNL SSPFPMDSVY KKFFPDEESP 480
481 PTPAKADKPC SSHPSSNFEP EKSDQLNNPE KITGQDNNTM AKEPTVLEHH DVIQEDPSLI 540
541 SDSVVANVGL ERRGGQEDDH HDSSGAISSQ DVPDGADCRT PDSPSCSQEQ SWDKESVNSN 600
601 NEERIKLLQD EVNSLRSKVA QLLSENARIL SDEKAKTSVA PKELNNEKVQ TEDADAPTSF 660
661 TPFIKYQKLE FKVDTFFAVG SPLGVFLALR NIRLGIGKGK DYWEEENAIE EMPACRRMFN 720
721 IFHPYDPVAY RVEPLVCKEY LPERPVIIPY HRGGKRLHIG LQDFREDFAA RSQRIMNHFD 780
781 SVRTRVLTIC QSKSADNLDE MEETDDEKDD RSYGSLMIER LTGTRDGRID HMLQEKTFEH 840
841 PYLQAIGAHT NYWRDQDTAL FIIKHLYREL PDGPNSPTES TEGDDSPKDS SRPHSWIDRR 900
901 EADYDDEELP LTFSDKQITR SFSAEAKKYL KKP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
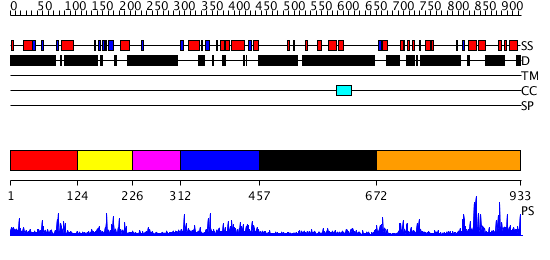
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..123] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [124..225] | 2.092979 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [226..311] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [312..456] | 2.111977 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [457..671] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [672..933] | 35.221849 | No description for PF02862.9 was found. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)