













| General Information: |
|
| Name(s) found: |
UBP12_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1116 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
DNA topological change
[IEA]
ubiquitin-dependent protein catabolic process [IEA] |
| Molecular Function: |
ubiquitin-specific protease activity
[ISS]
double-stranded DNA binding [IEA] ubiquitin thiolesterase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTMMTPPPVD QPEDEEMLVP NSDLVDGPAQ PMEVTQPETA ASTVENQPAE DPPTLKFTWT 60
61 IPNFSRQNTR KHYSDVFVVG GYKWRILIFP KGNNVDHLSM YLDVSDAASL PYGWSRYAQF 120
121 SLAVVNQIHT RYTVRKETQH QFNARESDWG FTSFMPLSEL YDPSRGYLVN DTVLVEAEVA 180
181 VRKVLDYWSY DSKKETGFVG LKNQGATCYM NSLLQTLYHI PYFRKAVYHM PTTENDAPTA 240
241 SIPLALQSLF YKLQYNDTSV ATKELTKSFG WDTYDSFMQH DVQELNRVLC EKLEDKMKGT 300
301 VVEGTIQQLF EGHHMNYIEC INVDFKSTRK ESFYDLQLDV KGCKDVYASF DKYVEVERLE 360
361 GDNKYHAEGH GLQDAKKGVL FIDFPPVLQL QLKRFEYDFM RDTMVKINDR YEFPLELDLD 420
421 REDGKYLSPD ADRSVRNLYT LHSVLVHSGG VHGGHYYAFI RPTLSDQWYK FDDERVTKED 480
481 LKRALEEQYG GEEELPQTNP GFNNNPPFKF TKYSNAYMLV YIRESDKDKI ICNVDEKDIA 540
541 EHLRVRLKKE QEEKEDKRRY KAQAHLYTII KVARDEDLKE QIGKDIYFDL VDHDKVRSFR 600
601 IQKQTPFQQF KEEVAKEFGV PVQLQRFWIW AKRQNHTYRP NRPLTPQEEL QPVGQIREAS 660
661 NKANTAELKL FLEVEHLDLR PIPPPEKSKE DILLFFKLYD PEKAVLSYAG RLMVKSSSKP 720
721 MDITGKLNEM VGFAPDEEIE LFEEIKFEPC VMCEHLDKKT SFRLCQIEDG DIICFQKPLV 780
781 NKEIECLYPA VPSFLEYVQN RQLVRFRALE KPKEDEFVLE LSKQHTYDDV VEKVAEKLGL 840
841 DDPSKLRLTS HNCYSQQPKP QPIKYRGVDH LSDMLVHYNQ TSDILYYEVL DIPLPELQGL 900
901 KTLKVAFHHA TKEEVVIHNI RLPKQSTVGD VINELKTKVE LSHPDAELRL LEVFYHKIYK 960
961 IFPSTERIEN INDQYWTLRA EEIPEEEKNI GPNDRLILVY HFAKETGQNQ QVQNFGEPFF1020
1021 LVIHEGETLE EIKNRIQKKL HVSDEDFAKW KFAFMSMGRP EYLQDTDVVY NRFQRRDVYG1080
1081 AFEQYLGLEH ADTTPKRAYA ANQNRHAYEK PVKIYN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
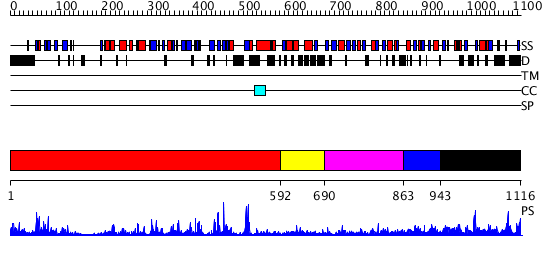
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..591] | 73.221849 | Crystal structure of HAUSP | |
| 2 | View Details | [592..689] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [690..862] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [863..942] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [943..1116] | 1.147993 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)