













| General Information: |
|
| Name(s) found: |
NAGS2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 613 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA]
|
| Biological Process: |
arginine biosynthetic process
[IEA][ISS]
metabolic process [IEA] cellular amino acid biosynthetic process [IEA] |
| Molecular Function: |
acetyl-CoA:L-glutamate N-acetyltransferase activity
[IEA][ISS]
N-acetyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERGALVGSS STSSYYVPYH FRQSKSNFSS FKPKNKLNPT QFRFNCSWFK PVSSITAAKC 60
61 NMFDYAVTAA GDVEAEHPVD DKQFVRWFRE AWPYLWAHRG CTFVVIISGE IIAGSSCDAI 120
121 LKDIAFLHHL GIRFVLVPGT QEQIDQLLAE RGREATYVGR YRVTDAASLQ AAKEAAGAIS 180
181 VMLEAKLSPG PSICNIRRHG DRSRLHDIGV RVDTGNFFAA KRRGVVDGVD FGATGEVKKI 240
241 DVDRICERLD GGSVVLLRNL GHSSSGEVLN CNTYEVATAC ALAIGADKLI CIMDGPILDE 300
301 SGHLIHFLTL QEADMLVRKR AQQSDIAANY VKAVGDGSMA YPEPPNNTNG NITSAQNGRA 360
361 VSFWGNGNHT PIFQNGVGFD NGNGLWPCEQ GFAIGGEERL SRLNGYLSEL AAAAFVCRGG 420
421 VKRVHLLDGT ISGVLLLELF KRDGMGTMVA SDVYEGTRDA RVEDLAGIRH IIKPLEESGI 480
481 LVRRTDEELL RALDSFVVVE REGQIIACAA LFPFFKDKCG EVAAIAVASD CRGQGQGDKL 540
541 LDYIEKKASS LGLEKLFLLT TRTADWFVRR GFQECSIEII PESRRQRINL SRNSKYYMKK 600
601 LIPDRSGISV MRI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
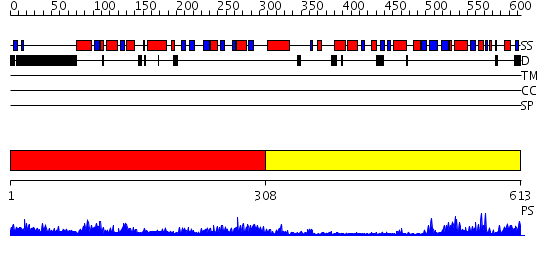
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..307] | 67.522879 | Acetylglutamate kinase from Thermotoga maritima complexed with its inhibitor arginine | |
| 2 | View Details | [308..613] | 6.69897 | The structure of Mycothiol Synthase in complex with des- AcetylMycothiol and CoenzymeA. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)