













| General Information: |
|
| Name(s) found: |
PCFS4_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 808 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] |
| Biological Process: |
positive regulation of flower development
[IMP]
mRNA polyadenylation [IMP] |
| Molecular Function: |
zinc ion binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSEKILNPR LVSINSTSRK GMSVELPQKP PPPPSLLDRF KALLNQREDE FGGGEEVLPP 60
61 SMDEIVQLYE VVLGELTFNS KPIITDLTII AGEQREHGEG IANAICTRIL EAPVEQKLPS 120
121 LYLLDSIVKN IGRDYGRYFS SRLPEVFCLA YRQAHPSLHP SMRHLFGTWS SVFPPPVLRK 180
181 IDMQLQLSSA ANQSSVGASE PSQPTRGIHV NPKYLRRLEP SAAENNLRGI NSSARVYGQN 240
241 SLGGYNDFED QLESPSSLSS TPDGFTRRSN DGANPSNQAF NYGMGRATSR DDEHMEWRRK 300
301 ENLGQGNDHE RPRALIDAYG VDTSKHVTIN KPIRDMNGMH SKMVTPWQNT EEEEFDWEDM 360
361 SPTLDRSRAG EFLRSSVPAL GSVRARPRVG NTSDFHLDSD IKNGVSHQLR ENWSLSQNYP 420
421 HTSNRVDTRA GKDLKVLASS VGLVSSNSEF GAPPFDSIQD VNSRFGRALP DGTWPHLSAR 480
481 GPNSLPVPSA HLHHLANPGN AMSNRLQGKP LYRPENQVSQ SHLNDMTQQN QMLVNYLPSS 540
541 SAMAPRPMQS LLTHVSHGYP PHGSTIRPSL SIQGGEAMHP LSSGVLSQIG ASNQPPGGAF 600
601 SGLIGSLMAQ GLISLNNQPA GQGPLGLEFD ADMLKIRNES AISALYGDLP RQCTTCGLRF 660
661 KCQEEHSKHM DWHVTKNRMS KNHKQNPSRK WFVSASMWLS GAEALGAEAV PGFLPTEPTT 720
721 EKKDDEDMAV PADEDQTSCA LCGEPFEDFY SDETEEWMYK GAVYMNAPEE STTDMDKSQL 780
781 GPIVHAKCRP ESNGGDMEEG SQRKKMRS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
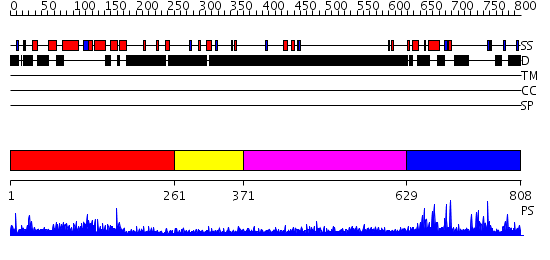
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..260] | 42.522879 | The RNA polymerase II CTD in mRNA processing: beta-turn recognition and beta-spiral model | |
| 2 | View Details | [261..370] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [371..628] | 4.286993 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [629..808] | 1.165973 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)