













| General Information: |
|
| Name(s) found: |
gi|42563399
[NCBI NR]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1056 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
transition metal ion binding
[IEA]
oxidoreductase activity [IEA] acid phosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEMEEGASGV GEKIKIGVCV MEKKVKCGSE VFSAPMGEIL DRLQSFGEFE ILHFGDKVIL 60
61 EDPIESWPIC DCLIAFHSSG YPLEKAQAYA ALRKPFLVNE LDPQYLLHDR RKVYEHLEMY 120
121 GIPVPRYACV NRKVPNQDLH YFVEEEDFVE VHGERFWKPF VEKPVNGDDH SIMIYYPSSA 180
181 GGGMKELFRK IGNRSSEFHP DVRRVRREGS YIYEEFMATG GTDVKVYTVG PEYAHAEARK 240
241 SPVVDGVVMR NTDGKEVRYP VLLTPAEKQM AREVCIAFRQ AVCGFDLLRS EGCSYVCDVN 300
301 GWSFVKNSYK YYDDAACVLR KMCLDAKAPH LSSTLPPTLP WKVNEPVQSN EGLTRQGSGI 360
361 IGTFGQSEEL RCVIAVVRHG DRTPKQKVKL KVTEEKLLNL MLKYNGGKPR AETKLKSAVQ 420
421 LQDLLDATRM LVPRTRPGRE SDSDAEDLEH AEKLRQVKAV LEEGGHFSGI YRKVQLKPLK 480
481 WVKIPKSDGD GEEERPVEAL MVLKYGGVLT HAGRKQAEEL GRYFRNNMYP GEGTGLLRLH 540
541 STYRHDLKIY SSDEGRVQMS AAAFAKGLLD LEGQLTPILV SLVSKDSSML DGLDNASIEM 600
601 EAAKARLNEI VTSGTKMIDD DQVSSEDFPW MTDGAGLPPN AHELLRELVK LTKNVTEQVR 660
661 LLAMDEDENL TEPYDIIPPY DQAKALGKTN IDSDRIASGL PCGSEGFLLM FARWIKLARD 720
721 LYNERKDRFD ITQIPDVYDS CKYDLLHNSH LDLKGLDELF KVAQLLADGV IPNEYGINPQ 780
781 QKLKIGSKIA RRLMGKILID LRNTREEALS VAELKESQEQ VLSLSASQRE DRNSQPKLFI 840
841 NSDELRRPGT GDKDEDDDKE TKYRLDPKYA NVKTPERHVR TRLYFTSESH IHSLMNVLRY 900
901 CNLDESLLGE ESLICQNALE RLCKTKELDY MSYIVLRLFE NTEVSLEDPK RFRIELTFSR 960
961 GADLSPLRNN DDEAETLLRE HTLPIMGPER LQEVGSCLSL ETMEKMVRPF AMPAEDFPPA1020
1021 STPVGFSGYF SKSAAVLERL VNLFHNYKNS SSNGRS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
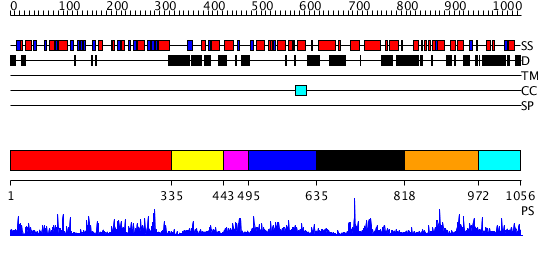
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..334] | 43.39794 | Crystal structure of a lysine biosynthesis enzyme, Lysx, from thermus thermophilus HB8 | |
| 2 | View Details | [335..442] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [443..494] | 1.219991 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [495..634] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [635..817] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [818..971] | 1.036974 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [972..1056] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)