













| General Information: |
|
| Name(s) found: |
FTSI2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 876 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
chloroplast envelope [IDA] |
| Biological Process: |
embryonic development ending in seed dormancy
[NAS]
|
| Molecular Function: |
ATPase activity
[ISS]
nucleotide binding [IEA] ATP binding [IEA] serine-type endopeptidase activity [IEA] nucleoside-triphosphatase activity [IEA] ATP-dependent peptidase activity [IEA] metalloendopeptidase activity [IEA] metallopeptidase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MACRFPLHSS SPSQFLSPEN RQRLPRNYPS ISCQNNSATN VVHEDGDDND KAKTNQVNLL 60
61 AIPITLTIIS ASLAKPSFAA AKVTERKRTQ KKPQEALTLE QLKAWSKDLP VVSNRIPYTD 120
121 ILSLKAEGKL KHVIKPPNLS LRQKAEPVLV VLEDSRVLRT VLPSLEGNKR FWEQWDELGI 180
181 DVQCVNAYTP PVKRPPVPSP YLGFLWKVPA YMLTWVKPKK ESKRAAELKR MREDFKRQRK 240
241 EEIETMKEER VMMEKTMKAQ KKQQERKKRK AVRKKKYEES LREARKNYRD MADMWARLAQ 300
301 DPNVATALGL VFFYIFYRVV VLNYRKQKKD YEDRLKIEKA EADERKKMRE LEREMEGIEE 360
361 EDEEVEEGTG EKNPYLQMAM QFMKSGARVR RASNKRLPEY LERGVDVKFT DVAGLGKIRL 420
421 ELEEIVKFFT HGEMYRRRGV KIPGGILLCG PPGVGKTLLA KAVAGEAGVN FFSISASQFV 480
481 EIYVGVGASR VRALYQEARE NAPSVVFIDE LDAVGRERGL IKGSGGQERD ATLNQLLVSL 540
541 DGFEGRGEVI TIASTNRPDI LDPALVRPGR FDRKIFIPKP GLIGRMEILQ VHARKKPMAE 600
601 DLDYMAVASM TDGMVGAELA NIVEIAAINM MRDGRTELTT DDLLQAAQIE ERGMLDRKDR 660
661 SLETWRQVAI NEAAMAVVAV NFPDMKNIEF LTINPRAGRE LGYVRVKMDH IKFKEGMLSR 720
721 QSILDHITVQ LAPRAADELW YGEDQLSTIW AETSDNARSA ARSLVLGGLS DKHHGLNNFW 780
781 VADRINDIDV EALRILNMCY ERAKEILGRN RTLMDEVVEK LVQKKSLTKQ EFFTLVELYG 840
841 SSKPMPPSIL ELRKIKRLEL EEMVLKLDMT TARNSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
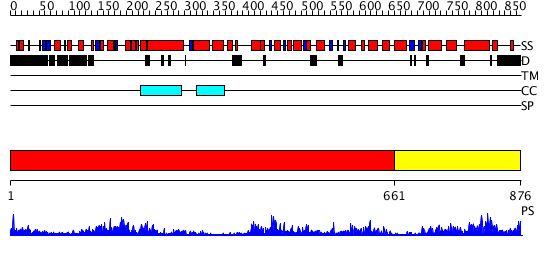
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..660] | 115.0 | VCP/p97 | |
| 2 | View Details | [661..876] | 106.0 | EDTA treated |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)