













| General Information: |
|
| Name(s) found: |
MCM4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 847 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
DNA replication initiation
[ISS]
DNA unwinding involved in replication [TAS] |
| Molecular Function: |
nucleotide binding
[IEA]
ATP binding [IEA][ISS] DNA binding [IEA][ISS] DNA-dependent ATPase activity [ISS] nucleoside-triphosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASDSSLGNT NDGPPSPGEN VSSPIENTYS SPAALHRRRR GRSSTPTQFA TPPPPPSRLA 60
61 SSNSTPPTSR PSAARSKGRN GHGGGGGGGG GGDPGTPMST DEPLPSSDDG EEDGGDDTTP 120
121 TFVWGTNISV QDVKSAIEMF VKHFREAREN SDDLFREGKY MVSIRKVIEI EGEWIDVDAF 180
181 DVFDYDPDLY NKMVRYPLEV LAIFDIVLMD IVSTINRLFE KHVQVRIFNL RTSTSMRNLN 240
241 PSDIEKMISL KGMIIRSSSI IPEIREAVFR CLVCGYFSDP IIVDRGKISE PPTCLKQECM 300
301 TKNSMTLVHN RCRFADKQIV RLQETPDEIP EGGTPHTVSL LLHDKLVDNG KPGDRIEVTG 360
361 IYRAMTVRVG PAHRTVKSVF KTYIDCLHIK KASKLRMSAE DPMDVDNSLR RVDEDVELDE 420
421 EKLRKFQELS KQPDIYERLS RSLAPNIWEL DDVKKGLLCQ LFGGNALNLA SGANFRGDIN 480
481 ILLVGDPGTS KSQLLQYIHK LSPRGIYTSG RGSSAVGLTA YVAKDPETGE TVLESGALVL 540
541 SDRGICCIDE FDKMSDSARS MLHEVMEQQT VSIAKAGIIA SLNARTSVLA CANPSGSRYN 600
601 PRLSVIENIH LPPTLLSRFD LIYLILDKPD EQTDRRLAKH IVALHFENAE SAQEEAIDIT 660
661 TLTTYVSYAR KNIHPKLSDE AAEELTRGYV ELRKAGKFAG SSKKVITATP RQIESLIRLS 720
721 EALARMRFSE WVEKHDVDEA FRLLRVAMQQ SATDHATGTI DMDLINTGVS ASERMRRDTF 780
781 ASSIRDIALE KMQIGGSSMR LSELLEELKK HGGNINTEIH LHDVRKAVAT LASEGFLVAE 840
841 GDRIKRV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
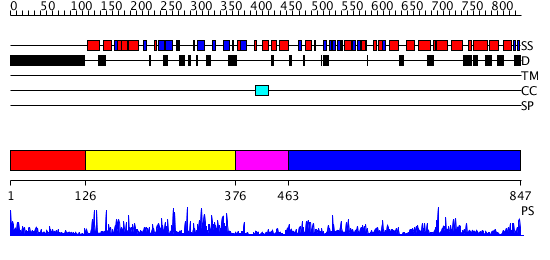
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..125] | 3.117997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [126..375] | 53.045757 | No description for 2vl6A was found. | |
| 3 | View Details | [376..462] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [463..847] | 50.0 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)