













| General Information: |
|
| Name(s) found: |
gi|2392701
[NCBI NR]
gi|2392700 [NCBI NR] gi|2392280 [NCBI NR] gi|2392279 [NCBI NR] gi|2392278 [NCBI NR] gi|2392277 [NCBI NR] gi|2392276 [NCBI NR] gi|2392275 [NCBI NR] gi|2392274 [NCBI NR] gi|2392273 [NCBI NR] gi|2392272 [NCBI NR] gi|2392271 [NCBI NR] gi|2392270 [NCBI NR] gi|2392269 [NCBI NR] gi|2392268 [NCBI NR] gi|2392267 [NCBI NR] |
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Rattus norvegicus |
| Length: | 624 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 GSMDQRQKLQ HWIHSCLRKA DKNKDNKMNF KELKDFLKEL NIQVDDGYAR KIFRECDHSQ 60
61 TDSLEDEEIE TFYKMLTQRA EIDRAFEEAA GSAETLSVER LVTFLQHQQR EEEAGPALAL 120
121 SLIERYEPSE TAKAQRQMTK DGFLMYLLSA DGNAFSLAHR RVYQDMDQPL SHYLVSSSHN 180
181 TYLLEDQLTG PSSTEAYIRA LCKGCRCLEL DCWDGPNQEP IIYHGYTFTS KILFCDVLRA 240
241 IRDYAFKASP YPVILSLENH CSLEQQRVMA RHLRAILGPI LLDQPLDGVT TSLPSPEQLK 300
301 GKILLKGKKL GGLLPAGGEN GSEATDVSDE VEAAEMEDEA VRSQVQHKPK EDKLKLVPEL 360
361 SDMIIYCKSV HFGGFSSPGT SGQAFYEMAS FSESRALRLL QESGNGFVRH NVSCLSRIYP 420
421 AGWRTDSSNY SPVEMWNGGC QIVALNFQTP GPEMDVYLGC FQDNGGCGYV LKPAFLRDPN 480
481 TTFNSRALTQ GPWWRPERLR VRIISGQQLP KVNKNKNSIV DPKVIVEIHG VGRDTGSRQT 540
541 AVITNNGFNP RWDMEFEFEV TVPDLALVRF MVEDYDSSSK NDFIGQSTIP WNSLKQGYRH 600
601 VHLLSKNGDQ HPSATLFVKI SIQD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
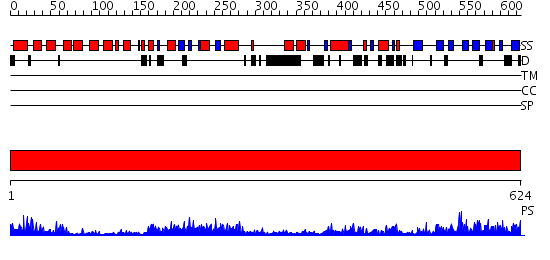
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..624] | 173.0 | Phosphoinositide-specific phospholipase C, isozyme D1 (PLC-D!); PI-specific phospholipase C isozyme D1 (PLC-D1), C-terminal domain; Phospholipase C isozyme D1 (PLC-D1) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)