













| General Information: |
|
| Name(s) found: |
IRC20 /
YLR247C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1556 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrion [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAVGALLAR EYNVTAEKCD FFLENGSFDS VIAALPALNQ EQETVTQKVS KNGKELINVA 60
61 SVNIEIPERI SLSNNGRQLF KFIVDIEILP CENDNEATLV VSSDSVSLFQ IGFKMENNSK 120
121 IAVDKQLPLI LDICAHKNQE RALRNQLENR KSLERKPSRK RRKKNSNVND PEKLLKSRIH 180
181 ELSLSYKDFE CSPVVFRYND SSLTWVLSFN LKLKFLNNKF NRFSVEANQI LDLTFSNRDE 240
241 NEFERYHKHS HIHSNFIQKQ FISQILEYSK DRLSKIKPFL PQSIPDLKVN LLPFQRESVE 300
301 WMLIKEGHGN SLSDTPTVID EVGLIDFMNE YYAYGYELIA RSPDEVGPSL LWNKLTGYIL 360
361 TTEDAAHLYN QYRKERLSGD YPVCAKGVLA EEMGLGKTIE ILSLILLNRR KLKDSEATFI 420
421 DDENRTITKT KTTLIICPNA ILKQWLEEIE LHANSLKWYT YRGYNEIMKD CKTVDEAVQQ 480
481 LCQYDIIVTS YNIIATEVHH AEFNRSIRSR RLKSPKYDYS SPLALMQFYR IILDEVQMLR 540
541 SSSTYSAKCT SLLHRIHTWG VSGTPIQNIY NFRMIMSYLK LHPFCDEVDF IRTLQEEIKL 600
601 RNEAKDYTSN DFVCQLKGVR FSIKDCMNIF YRYDLCIRHS KANVASQIHI PRQHNFIIPL 660
661 EFAPIEWDNY LNLWNNFLEL SGYNSDGSGS PRVSNAFLNE WLSRLRYICC HALFPEILST 720
721 RQKRLHGHLS RISNIDDILI SMRMDAFDSL IGYYRERFHL SIKQAQYELE ISNTPAKALE 780
781 SFIKIRDDLM IHIRQKFNVE DPFDKSLNLS EDEDEHMDER FGEKETSSGD ESDREINGAK 840
841 NHDNHNNDGM LSNHLKKKGL RAMMNLLHDC YFFLGSVYYN LGTRKLEEAD DKHRKEKTEE 900
901 VVYSDVFPKN ELEEIEENRL LEQENYANAE ILRKSILSSE ARKVDMTIKM ARTKFAPMTS 960
961 NIPLRLINIE FDHKNDYSSN LAVSRCFKSL SKLIEGLNEQ TKNFNELLDE LLIIIYEPVH1020
1021 RTEDDDSTNK IIGNEEYSTS IDSQDKIFSL LGCLEIILQN RDNILTSESE VKIPKHLVPE1080
1081 GSIISKYQKQ LLNSLRLISG TPLRTVFDEL KNSRIVRRIS SSNESESTIQ NFEDYLLQYE1140
1141 VESKSLFKYN KQVRESLKIL GSIYNAKTEY YSQLQRISDS LVSLHSLSAP QLSHLIRTIN1200
1201 KSLGGTLDAK INNIESRLIY LKNLSRLKDT LNDNQILSCS ICLGEVEIGA IIKCGHYFCK1260
1261 SCILTWLRAH SKCPICKGFC SISEVYNFKF KNSTEKREKE IQEPRREGAD SSQDNSNENS1320
1321 IISNMSEVEK LFGNKYEQFH QINEVHQIHI KESFGAKIDF VIKLISYLRL KSEQENADPP1380
1381 QVILYSQKTE YLKVIGKVLK LYHIEHLACL SNTANVGETI NNFKRQPSVT CLLLNVKTLG1440
1441 AGLNLINAKH IFLLDPILNN SDELQAMGRN NRIGQDEETF VWNFMIRNTV EENILRYKCI1500
1501 LEERKRKEKS KKGDKYDEAQ DETDNEESDD AKFEISVVDQ EVSNEHLWNC FFHGSD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
ARC1 ATG14 CAT8 CCC2 HEK2 IRC20 MCA1 MES1 MGA2 NCS2 NHA1 PET127 PHO89 PHO91 YKL050C YLR363W-A |
| View Details | Ho Y, et al. (2002) |
|
CIN5 EXO70 HHF1, HHF2 HTA1 IRC20 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
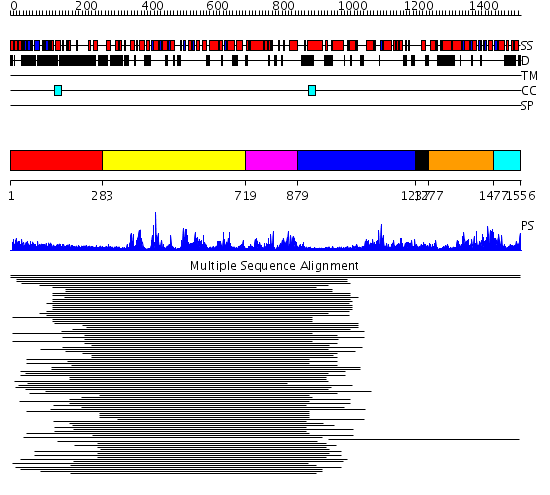
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..282] | 1.193996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [283..718] | 8.638272 | SNF2 family N-terminal domain No confident structure predictions are available. | |
| 3 | View Details | [719..878] | 2.253996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [879..1236] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1237..1276] | 7.568636 | Zinc finger, C3HC4 type (RING finger) No confident structure predictions are available. | |
| 6 | View Details | [1277..1476] | 4.522879 | Putative DEAD box RNA helicase | |
| 7 | View Details | [1477..1556] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)