













| General Information: |
|
| Name(s) found: |
MEF2 /
YJL102W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 819 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
translational elongation
[ISS |
| Molecular Function: |
translation elongation factor activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWKWNVRRWA GARVNISKNR LSVINVGSRY LSTARSPLSK VRNIGIIAHI DAGKTTTTER 60
61 MLYYAGISKH IGDVDTGDTI TDFLEQERSR GITIQSAAIS FPWRNTFAIN LIDTPGHIDF 120
121 TFEVIRALKV IDSCVVILDA VAGVEAQTEK VWKQSKSKPK ICFINKMDRM GASFNHTVND 180
181 LINKFMRGTT TKPVLVNIPY YRKQPTSNDY VFQGVIDVVN GKRLTWNPEN PDEIIVDELD 240
241 GTSLEQCNRC RESMIETLTE YDEDLVQHFL EEAEGDYSKV SAQFLNASIR KLTMKNMIVP 300
301 VLCGASFKNI GVQPLLDAIV NYLPSPIEAE LPELNDKTVP MKYDPKVGCL VNNNKNLCIA 360
361 LAFKVITDPI RGKQIFIRIY SGTLNSGNTV YNSTTGEKFK LGKLLIPHAG TSQPVNILTA 420
421 GQIGLLTGST VENNISTGDT LITHSSKKDG LKSLDKKKEL TLKINSIFIP PPVFGVSIEP 480
481 RTLSNKKSME EALNTLITED PSLSISQNDE TGQTVLNGMG ELHLEIAKDR LVNDLKADVE 540
541 FGQLMVSYKE TINSETNIET YESDDGYRFS LSLLPNSDAL PNCLAYPLGV NENFLIMEKN 600
601 GNWDKEWKYQ VSFESILNSI IASCIVGLQR GGKIANFPLY ACSIKINSDW SVPPDIETPQ 660
661 EILKITRNLI FKALNDLKPE KYNLLEPIMN LDLTIPQSDV GTVLQDLTGA RKAQILSIED 720
721 ESSVSNSGAS TCNSPENSNR IYIPSDAVTT LHATKDKKNT QETSSNVKKI IKAKVPLREI 780
781 TTYTNKLRSL SQGRGEFNIE YSDMEKVTND RLQSILHDL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
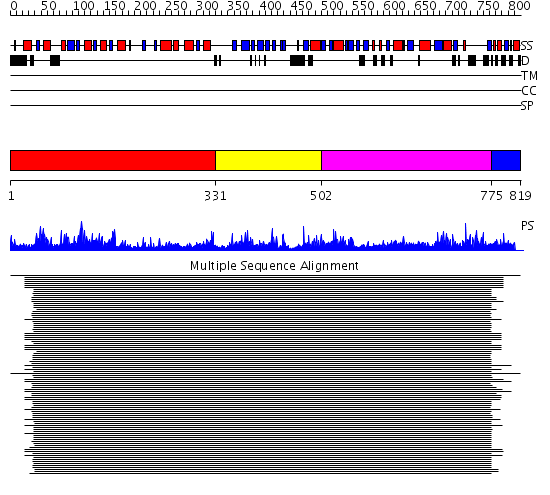
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..330] | 2060.0 | Elongation factor G (EF-G), domain II; Elongation factor G (EF-G), N-terminal (G) domain; Elongation factor G (EF-G), domain IV; Elongation factor G (EF-G) | |
| 2 | View Details | [331..501] | 2060.0 | Elongation factor G (EF-G), domain II; Elongation factor G (EF-G), N-terminal (G) domain; Elongation factor G (EF-G), domain IV; Elongation factor G (EF-G) | |
| 3 | View Details | [502..774] | 2060.0 | Elongation factor G (EF-G), domain II; Elongation factor G (EF-G), N-terminal (G) domain; Elongation factor G (EF-G), domain IV; Elongation factor G (EF-G) | |
| 4 | View Details | [775..819] | 8.33 | Elongation factor G (EF-G), domain II; Elongation factor G (EF-G), N-terminal (G) domain; Elongation factor G (EF-G), domain IV; Elongation factor G (EF-G) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)