













| General Information: |
|
| Name(s) found: |
XKS1 /
YGR194C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 600 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
xylulose catabolic process
[IDA |
| Molecular Function: |
xylulokinase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLCSVIQRQT REVSNTMSLD SYYLGFDLST QQLKCLAINQ DLKIVHSETV EFEKDLPHYH 60
61 TKKGVYIHGD TIECPVAMWL EALDLVLSKY REAKFPLNKV MAVSGSCQQH GSVYWSSQAE 120
121 SLLEQLNKKP EKDLLHYVSS VAFARQTAPN WQDHSTAKQC QEFEECIGGP EKMAQLTGSR 180
181 AHFRFTGPQI LKIAQLEPEA YEKTKTISLV SNFLTSILVG HLVELEEADA CGMNLYDIRE 240
241 RKFSDELLHL IDSSSKDKTI RQKLMRAPMK NLIAGTICKY FIEKYGFNTN CKVSPMTGDN 300
301 LATICSLPLR KNDVLVSLGT STTVLLVTDK YHPSPNYHLF IHPTLPNHYM GMICYCNGSL 360
361 ARERIRDELN KERENNYEKT NDWTLFNQAV LDDSESSENE LGVYFPLGEI VPSVKAINKR 420
421 VIFNPKTGMI EREVAKFKDK RHDAKNIVES QALSCRVRIS PLLSDSNASS QQRLNEDTIV 480
481 KFDYDESPLR DYLNKRPERT FFVGGASKND AIVKKFAQVI GATKGNFRLE TPNSCALGGC 540
541 YKAMWSLLYD SNKIAVPFDK FLNDNFPWHV MESISDVDNE NWDRYNSKIV PLSELEKTLI 600
601 |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |
|
PRO1 XKS1 |
| View Details | Gavin AC, et al. (2002) |

|
CCT4 CHC1 COG8 GCD1 GCD11 GCD2 GCD6 GCD7 GCN3 GUT1 IST1 PDC1 PRO1 RRP40 RRP46 SUI2 SUI3 XKS1 |
| View Details | Qiu et al. (2008) |

|
GLK1 XKS1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
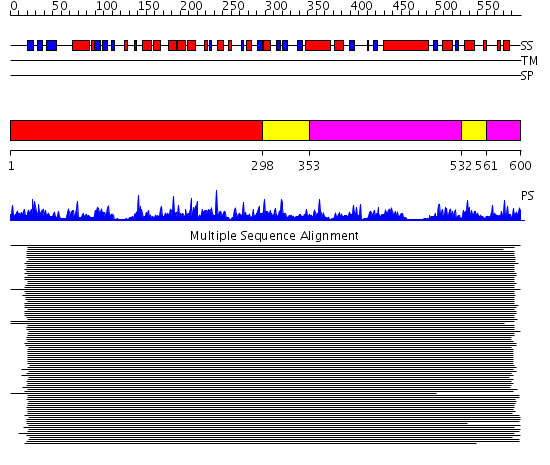
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..297] | 141.0 | Glycerol kinase | |
| 2 | View Details | [298..352] [532..560] |
141.0 | Glycerol kinase | |
| 3 | View Details | [353..531] [561..600] |
141.0 | Glycerol kinase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)