













| General Information: |
|
| Name(s) found: |
RPO41 /
YFL036W
[SGD]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1351 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IDA mitochondrion [IDA mitochondrial nucleoid [IDA |
| Biological Process: |
transcription from mitochondrial promoter
[IMP mitochondrial genome maintenance [IMP |
| Molecular Function: |
DNA-directed RNA polymerase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRPAYKSLV KTSLLQRRLI SSKGSKLFKP SPDSTSTILI SEDPLVTGSS PTSSTTSGII 60
61 SSNDFPLFNK NRKDAKSSIS YQWKNPSELE FDPFNKSHAS AVTSMTRTRD VMQLWSLLEA 120
121 CLQSNLMKRA FSILESLYLV PEHKQRFIED YNMYLNSFSK NDPNFPILKM NEKLTNDLET 180
181 SFKDVNYNDK TLAIMIHHAL NFHSTTSSML LKPIISAYLK MSVNGIREIF SCLDILTISD 240
241 LNILMNDLKV ITPSQLPNSV RPILESLTLS PTPVNNIENE EGLNKVEAEN DSKLHKASNA 300
301 SSDSIKKPSL DPLREVSFHG STEVLSKDAE KLIAVDTIGM RVIRHTLLGL SLTPEQKEQI 360
361 SKFKFDANDN VLKMKPTKND DNNNSINFFE IYNSLPTLEE KKAFESALNI FNQDRQKVLE 420
421 NRATEAARER WKHDFEEAKA RGDISIEKNL NVKLWKWYNE MLPLVKEEIN HCRSLLSEKL 480
481 SDKKGLNKVD TNRLGYGPYL TLIDPGKMCV ITILELLKLN STGGVIEGMR TARAVISVGK 540
541 AIEMEFRSEQ VLKSESQAFR DVNKKSPEFK KLVQNAKSVF RSSQIEQSKI LWPQSIRARI 600
601 GSVLISMLIQ VAKVSVQGVD PVTKAKVHGE APAFAHGYQY HNGSKLGVLK IHKTLIRQLN 660
661 GERLIASVQP QLLPMLVEPK PWVNWRSGGY HYTQSTLLRT KDSPEQVAYL KAASDNGDID 720
721 RVYDGLNVLG RTPWTVNRKV FDVVSQVWNK GEGFLDIPGA QDEMVLPPAP PKNSDPSILR 780
781 AWKLQVKTIA NKFSSDRSNR CDTNYKLEIA RAFLGEKLYF PHNLDFRGRA YPLSPHFNHL 840
841 GNDMSRGLLI FWHGKKLGPS GLKWLKIHLS NLFGFDKLPL KDRVAFTESH LQDIKDSAEN 900
901 PLTGDRWWTT ADKPWQALAT CFELNEVMKM DNPEEFISHQ PVHQDGTCNG LQHYAALGGD 960
961 VEGATQVNLV PSDKPQDVYA HVARLVQKRL EIAAEKGDEN AKILKDKITR KVVKQTVMTN1020
1021 VYGVTYVGAT FQIAKQLSPI FDDRKESLDF SKYLTKHVFS AIRELFHSAH LIQDWLGESA1080
1081 KRISKSIRLD VDEKSFKNGN KPDFMSSVIW TTPLGLPIVQ PYREESKKQV ETNLQTVFIS1140
1141 DPFAVNPVNA RRQKAGLPPN FIHSLDASHM LLSAAECGKQ GLDFASVHDS YWTHASDIDT1200
1201 MNVVLREQFI KLHEVDLVLR LKEEFDQRYK NYVKIGKLKR STDLAQKIIR IRKDLSRKLG1260
1261 RSTTLADEIY FEKKRQELLN SPLIEDRNVG EKMVTTVSLF EDITDLDALE LENGGDENSG1320
1321 MSVLLPLRLP EIPPKGDFDV TVLRNSQYFF S |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
ACO1 ILV5 MTF1 RIM1 RPO41 YHM2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
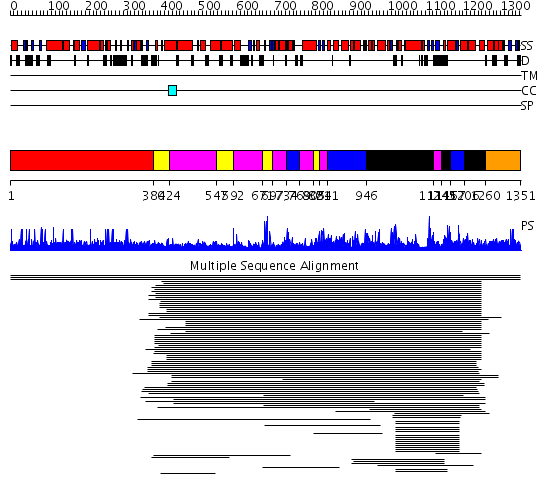
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..379] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [380..423] [547..591] [671..696] [805..820] |
1703.0103 | T7 RNA polymerase | |
| 3 | View Details | [424..546] [592..670] [697..733] [769..804] [821..840] [1124..1144] |
1703.0103 | T7 RNA polymerase | |
| 4 | View Details | [734..768] [841..945] [1167..1205] |
1703.0103 | T7 RNA polymerase | |
| 5 | View Details | [946..1123] [1145..1166] [1206..1259] |
1703.0103 | T7 RNA polymerase | |
| 6 | View Details | [1260..1351] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)