













| General Information: |
|
| Name(s) found: |
PTIBC_ECOLI
[Swiss-Prot]
PTDA_ECOLI [Swiss-Prot] |
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 485 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKNYAALAR SVIAALGGVD NISAVTHCMT RLRFVIKDDA LIDSPTLKTI PGVLGVVRSD 60
61 NQCQVIIGNT VSQAFQEVVS LLPGDMQPAQ PVGKPKLTLR RIGAGILDAL IGTMSPLIPA 120
121 IIGGSMVKLL AMILEMSGVL TKGSPTLTIL NVIGDGAFFF LPLMVAASAA IKFKTNMSLA 180
181 IAIAGVLVHP SFIELMAKAA QGEHVEFALI PVTAVKYTYT VIPALVMTWC LSYIERWVDS 240
241 ITPAVTKNFL KPMLIVLIAA PLAILLIGPI GIWIGSAISA LVYTIHGYLG WLSVAIMGAL 300
301 WPLLVMTGMH RVFTPTIIQT IAETGKEGMV MPSEIGANLS LGGSSLAVAW KTKNPELRQT 360
361 ALAAAASAIM AGISEPALYG VAIRLKRPLI ASLISGFICG AVAGMAGLAS HSMAAPGLFT 420
421 SVQFFDPANP MSIVWVFAVM ALAVVLSFIL TLLLGFEDIP VEEAAAQARK YQSVQPTVAK 480
481 EVSLN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
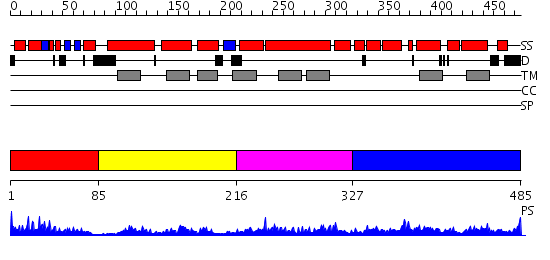
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | 11.30103 | Glucose permease domain IIB | |
| 2 | View Details | [85..215] | 3.582996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [216..326] | 1.599997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [327..485] | 6.597997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|