













| General Information: |
|
| Name(s) found: |
ARGT_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 260 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKKSILALSL LVGLSTAASS YAALPETVRI GTDTTYAPFS SKDAKGDFVG FDIDLGNEMC 60
61 KRMQVKCTWV ASDFDALIPS LKAKKIDAII SSLSITDKRQ QEIAFSDKLY AADSRLIAAK 120
121 GSPIQPTLDS LKGKHVGVLQ GSTQEAYANE TWRSKGVDVV AYANQDLVYS DLAAGRLDAA 180
181 LQDEVAASEG FLKQPAGKDF AFAGSSVKDK KYFGDGTGVG LRKDDAELTA AFNKALGELR 240
241 QDGTYDKMAK KYFDFNVYGD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
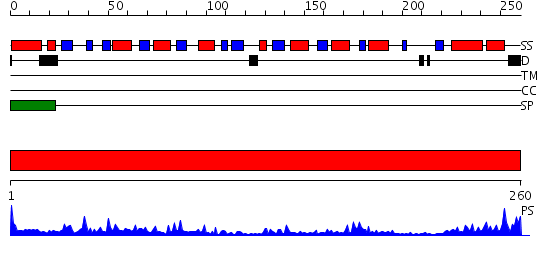
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..260] | 57.045757 | Lysine-,arginine-,ornithine-binding (LAO) protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|