













| General Information: |
|
| Name(s) found: |
Y5483_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 342 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] |
| Biological Process: |
circadian regulation of gene expression
[IMP]
photomorphogenesis [IMP] |
| Molecular Function: |
GTPase activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKFWRERERE NKEQILAPLC GQVRVLVVGD SGVGKTSLVH LINKGSSIVR PPQTIGCTVG 60
61 VKHITYGSPA SSSSSIQGDS ERDFFVELWD VSGHERYKDC RSLFYSQING VIFVHDLSQR 120
121 RTKTSLQKWA SEVAATGTFS APLPSGGPGG LPVPYIVVGN KADIAAKEGT KGSSGNLVDA 180
181 ARHWVEKQGL LPSSSEDLPL FESFPGNGGL IAAAKETRYD KEALNKFFRM LIRRRYFSDE 240
241 LPAASPWSIS PVPTSSSQRL DEITSDDDQF YKRTSFHGDP YKYNNTIPPL PAQRNLTPPP 300
301 TLYPQQPVST PDNYTIPRYS LSSVQETTNN GSARSKRMDI NV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
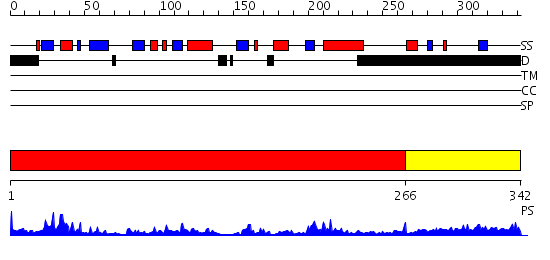
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..265] | 47.30103 | Structure of doubly prenylated Ypt1:GDI complex | |
| 2 | View Details | [266..342] | 2.330986 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)