













| General Information: |
|
| Name(s) found: |
prp2 /
SPBC146.07
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 517 amino acids |
Gene Ontology: |
|
| Cellular Component: |
commitment complex
[IDA nucleus [IDA U2 snRNP [TAS |
| Biological Process: |
RNA splicing, via transesterification reactions
[ISS generation of catalytic spliceosome for first transesterification step [IDA nuclear mRNA branch site recognition [ISS nuclear mRNA splicing, via spliceosome [IMP |
| Molecular Function: |
snoRNA binding
[TAS mRNA binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLSSRLSSG SSRIPKRHRD YRDEEPRRER GSGGIGREDP RGHYGSERPR RRRRDESDFR 60
61 RHRESRERSY REDERPRRER RYDDYEPRSL RYSSVGRSRS PPPSRERSVR SIEQELEQLR 120
121 DVTPINQWKR KRSLWDIKPP GYELVTADQA KMSGVFPLPG APRAAVTDPE KLLEFARSAE 180
181 GSIIAPPPPL QPGASRQARR LVVTGIPNEF VEDAFVSFIE DLFISTTYHK PETKHFSSVN 240
241 VCKEENFAIL EVATPEDATF LWGLQSESYS NDVFLKFQRI QNYIVPQITP EVSQKRSDDY 300
301 AKNDVLDSKD KIYISNLPLN LGEDQVVELL KPFGDLLSFQ LIKNIADGSS KGFCFCEFKN 360
361 PSDAEVAISG LDGKDTYGNK LHAQFACVGL NQAMIDKSNG MAILTELAKA SSQSIPTRVL 420
421 QLHNLITGDE IMDVQEYEDI YESVKTQFSN YGPLIDIKIP RSIGTRNSGL GTGKVFVRYS 480
481 DIRSAEVAME EMKGCKFNDR TIVIAFYGED CYKANAW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
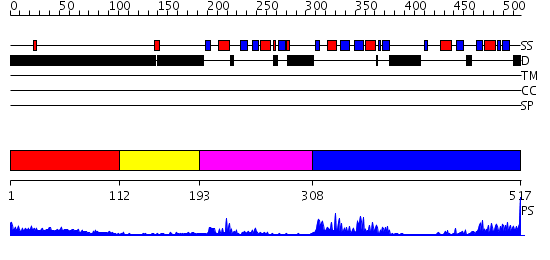
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | 5.128997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [112..192] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [193..307] | 29.522879 | Hu antigen D (Hud) | |
| 4 | View Details | [308..517] | 29.154902 | Poly(A)-binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)