













| General Information: |
|
| Name(s) found: |
tup11 /
SPAC18B11.10
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 614 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [IC cytosol [IDA protein complex [RCA |
| Biological Process: |
cellular response to stress
[IEP negative regulation of transcription by glucose [IMP chromatin silencing [TAS negative regulation of gene-specific transcription from RNA polymerase II promoter [IEP |
| Molecular Function: |
protein binding
[IPI histone binding [IDA transcription corepressor activity [IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASVEDATKV QEMLDALKAE YNALAHHSFA SKARGNDYES SMIQSQIQEI EAFRKTVDDM 60
61 YEKQKSIRET YEKDINKLKR ELEELGVEAN TASYRNRGER SELAASNNQV THIDQEHPSQ 120
121 TKSTSQPPSN HLPAFQQIPP IHQSAYPQNN VAEVLMPPIP PSVEASSGQN FNQGIASQNP 180
181 AISTSNLPST TPLYIPPVNY GANQVSQQPN PQLPGVSNYY NPSATSKPAV NVQPPRIPTK 240
241 ATPSAEPSMT ASANAGSISQ AGPDGEYQGR EQIAPVSDTE AARKTTSQSW YVTYNPACKR 300
301 VFNINLVHTL EHPSVVCCVK FSNNGKYLAT GCNQAANVFD VQTGKKLFTL HEESPDPSRD 360
361 LYVRTIAFSP DGKYLVTGTE DRQIKLWDLS TQKVRYVFSG HEQDIYSLDF SHNGRFIVSG 420
421 SGDRTARLWD VETGQCILKL EIENGVTAIA ISPNDQFIAV GSLDQIIRVW SVSGTLVERL 480
481 EGHKESVYSI AFSPDSSILL SGSLDKTIKV WELQATRSVG LSAIKPEGIC KATYTGHTDF 540
541 VLSVAVSPDS RWGLSGSKDR SMQFWDLQTG QSYLTCQGHK NSVISVCFSP DGRQFASGSG 600
601 DLRARIWSID PASP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
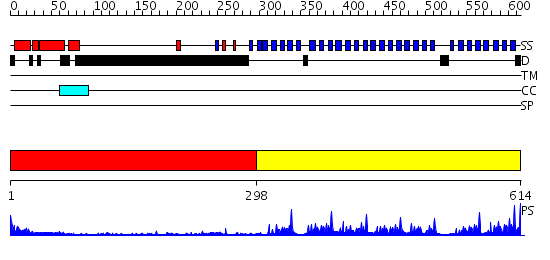
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..297] | 1.35 | Fibrinogen | |
| 2 | View Details | [298..614] | 63.221849 | Tup1, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)