













| General Information: |
|
| Name(s) found: |
FBpp0312027
[FlyBase]
Klp10A-PC / FBpp0073335 [FlyBase] Klp10A-PB / FBpp0073334 [FlyBase] Klp10A-PD / FBpp0073333 [FlyBase] Klp10A-PE / FBpp0073332 [FlyBase] Klp10A-PA / FBpp0073331 [FlyBase] |
| Description(s) found:
Found 81 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 805 amino acids |
Gene Ontology: |
|
| Cellular Component: |
kinetochore microtubule
[IDA]
spindle pole [IDA] kinesin complex [ISS] |
| Biological Process: |
spindle organization
[IMP]
mitotic spindle organization [IMP] mitotic sister chromatid segregation [IMP] mitotic anaphase A [IDA] microtubule-based movement [IEA] |
| Molecular Function: |
ATP binding
[IEA]
SUMO binding [IPI] microtubule motor activity [IEA] motor activity [IDA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDMITVGQSV KIKRTDGRVH MAVVAVINQS GKCITVEWYE RGETKGKEVE LDAILTLNPE 60
61 LMQDTVEQHA APEPKKQATA PMNLSRNPTQ SAIGGNLTSR MTMAGNMLNK IQESQSIPNP 120
121 IVSSNSVNTN SNSNTTAGGG GGTTTSTTTG LQRPRYSQAA TGQQQTRIAS AVPNNTLPNP 180
181 SAAASAGPAA QGVATAATTQ GAGGASTRRS HALKEVERLK ENREKRRARQ AEMKEEKVAL 240
241 MNQDPGNPNW ETAQMIREYQ STLEFVPLLD GQAVDDHQIT VCVRKRPISR KEVNRKEIDV 300
301 ISVPRKDMLI VHEPRSKVDL TKFLENHKFR FDYAFNDTCD NAMVYKYTAK PLVKTIFEGG 360
361 MATCFAYGQT GSGKTHTMGG EFNGKVQDCK NGIYAMAAKD VFVTLNMPRY RAMNLVVSAS 420
421 FFEIYSGKVF DLLSDKQKLR VLEDGKQQVQ VVGLTEKVVD GVEEVLKLIQ HGNAARTSGQ 480
481 TSANSNSSRS HAVFQIVLRP QGSTKIHGKF SFIDLAGNER GVDTSSADRQ TRMEGAEINK 540
541 SLLALKECIR ALGKQSAHLP FRVSKLTQVL RDSFIGEKSK TCMIAMISPG LSSCEHTLNT 600
601 LRYADRVKEL VVKDIVEVCP GGDTEPIEIT DDEEEEELNM VHPHSHQLHP NSHAPASQSN 660
661 NQRAPASHHS GAVIHNNNNN NNKNGNAGNM DLAMLSSLSE HEMSDELIVQ HQAIDDLQQT 720
721 EEMVVEYHRT VNATLETFLA ESKALYNLTN YVDYDQDSYC KRGESMFSQL LDIAIQCRDM 780
781 MAEYRAKLAK EEMLSCSFNS PNGKR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
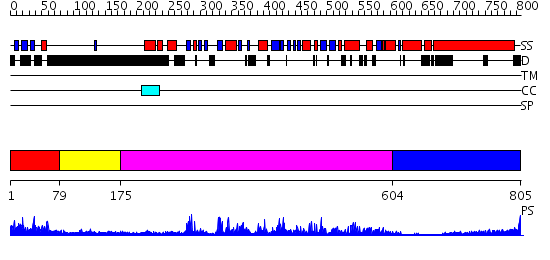
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..78] | 0.954 | No description for 2d9tA was found. | |
| 2 | View Details | [79..174] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [175..603] | 101.0 | No description for 2gryA was found. | |
| 4 | View Details | [604..805] | 7.0 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)