













| General Information: |
|
| Name(s) found: |
mof-PA /
FBpp0070794
[FlyBase]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 827 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
X chromosome [TAS] chromatin [IEA] X chromosome located dosage compensation complex, transcription activating [NAS][TAS] |
| Biological Process: |
dosage compensation
[IGI][IMP][NAS][TAS]
peptidyl-lysine acetylation [TAS] chromatin assembly or disassembly [IEA] histone acetylation [NAS][TAS] dosage compensation, by hyperactivation of X chromosome [NAS] regulation of transcription [IEA] |
| Molecular Function: |
histone acetyltransferase activity
[ISS][NAS][TAS]
lysine N-acetyltransferase activity [TAS] protein binding [TAS] mRNA binding [TAS] histone acetyltransferase activity (H4-K16 specific) [TAS] chromatin binding [IDA][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEAELEQTP SAGHVQEQPI EEEHEPEQEP TDAYTIGGPP RTPVEDAAAE LSASLDVSGS 60
61 DQSAEQSLDL SGVQAEAAAE SEPPAKRQHR DISPISEDST PASSTSTSST RSSSSSRYDD 120
121 VSEAEEAPPE PEPEQPQQQQ QEEKKEDGQD QVKSPGPVEL EAQEPAQPQK QKEVVDQEIE 180
181 TEDEPSSDTV ICVADINPYG SGSNIDDFVM DPDAPPNAII TEVVTIPAPL HLKGTQQLGL 240
241 PLAAPPPPPP PPAAEQVPET PASPTDDGEE PPAVYLSPYI RSRYMQESTP GLPTRLAPRD 300
301 PRQRNMPPPA VVLPIQTVLS ANVEAISDDS SETSSSDDDE EEEEDEDDAL TMEHDNTSRE 360
361 TVITTGDPLM QKIDISENPD KIYFIRREDG TVHRGQVLQS RTTENAAAPD EYYVHYVGLN 420
421 RRLDGWVGRH RISDNADDLG GITVLPAPPL APDQPSTSRE MLAQQAAAAA AASSERQKRA 480
481 ANKDYYLSYC ENSRYDYSDR KMTRYQKRRY DEINHVQKSH AELTATQAAL EKEHESITKI 540
541 KYIDKLQFGN YEIDTWYFSP FPEEYGKART LYVCEYCLKY MRFRSSYAYH LHECDRRRPP 600
601 GREIYRKGNI SIYEVNGKEE SLYCQLLCLM AKLFLDHKVL YFDMDPFLFY ILCETDKEGS 660
661 HIVGYFSKEK KSLENYNVAC ILVLPPHQRK GFGKLLIAFS YELSRKEGVI GSPEKPLSDL 720
721 GRLSYRSYWA YTLLELMKTR CAPEQITIKE LSEMSGITHD DIIYTLQSMK MIKYWKGQNV 780
781 ICVTSKTIQD HLQLPQFKQP KLTIDTDYLV WSPQTAAAVV RAPGNSG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
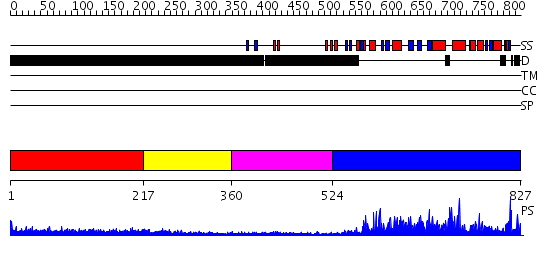
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..216] | 3.237995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [217..359] | 1.244996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [360..523] | 3.21 | Solution Structure of the Tudor Domain from Mouse Hypothetical Protein Homologous to Histone Acetyltransferase | |
| 4 | View Details | [524..827] | 135.0 | No description for 2givA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)