













| General Information: |
|
| Name(s) found: |
osk-PA /
FBpp0081435
[FlyBase]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 606 amino acids |
Gene Ontology: |
|
| Cellular Component: |
P granule
[IDA][TAS]
pole plasm [TAS] |
| Biological Process: |
germ cell development
[IMP]
positive regulation of translation [NAS] anterior/posterior axis specification, embryo [TAS] pole cell fate determination [NAS] pole cell formation [TAS] P granule organization [IMP][TAS] oocyte microtubule cytoskeleton polarization [IMP] regulation of mRNA stability [IMP] visual learning [IMP] pole cell development [TAS] pole plasm assembly [NAS][TAS] posterior abdomen determination [IMP] visual behavior [IMP] oocyte anterior/posterior axis specification [NAS] pole plasm mRNA localization [TAS] long-term memory [IMP][TAS] pole plasm protein localization [NAS][TAS] oogenesis [IMP] regulation of oskar mRNA translation [TAS] maintenance of pole plasm mRNA location [NAS] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAVTSEFPS KPISYTSTNT SAKTYYLKSV KKRVTTCFQQ LRDKLQSSGS FRKSSSSCLN 60
61 QIFVRSDFSA CGERFRKIFK SARKTELPEL WKVPLVAHEL TSRQSSQQLQ VVARLFSSTQ 120
121 ISTKEITYNS NSNTSENNMT IIESNYISVR EEYPDIDSEV RAILLSHAQN GITISSIKSE 180
181 YRKLTGNPFP LHDNVTDFLL TIPNVTAECS ESGKRIFNLK ASLKNGHLLD MVLNQKERTS 240
241 DYSSGAPSLE NIPRAPPRYW KNPFKRRALS QLNTSPRTVP KITDEKTKDI ATRPVSLHQM 300
301 ANEAAESNWC YQDNWKHLNN FYQQASVNAP KMPVPINIYS PDAPEEPINL APPGHQPSCR 360
361 TQSQKTEPTE NRHLGIFVHP FNGMNIMKRR HEMTPTPTIL TSGTYNDSLL TINSDYDAYL 420
421 LDFPLMGDDF MLYLARMELK CRFRRHERVL QSGLCVSGLT INGARNRLKR VQLPEGTQII 480
481 VNIGSVDIMR GKPLVQIEHD FRLLIKEMHN MRLVPILTNL APLGNYCHDK VLCDKIYRFN 540
541 KFIRSECCHL KVIDIHSCLI NERGVVRFDC FQASPRQVTG SKEPYLFWNK IGRQRVLQVI 600
601 ETSLEY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
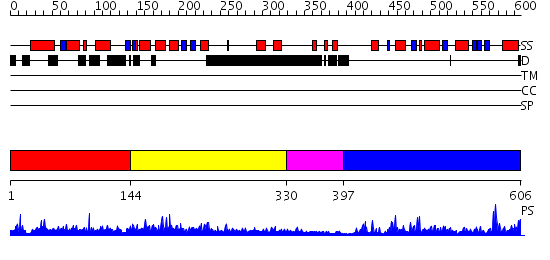
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [144..329] | 5.064981 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [330..396] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [397..606] | 41.045757 | Thioesterase I, TAP |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)