













| General Information: |
|
| Name(s) found: |
casp-PB /
FBpp0086398
[FlyBase]
casp-PA / FBpp0086397 [FlyBase] |
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 695 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
defense response to Gram-negative bacterium
[IMP]
|
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSENKDEALT SFQSITGIDD VGEAFSHLEA ADWNLVEALS RVIPHEDAPL TSPHPILVPE 60
61 PPARNAVEST NGFRPPGFDE MPGTSNASGF FAAALQNQNQ NLNLNQPHIQ QRFPTSQLLA 120
121 QLTSSINLDP ASSQNNNQNV ITFNIHFNQQ LYQLRLPSEA TVEQLKRKIF AETSVPVCRQ 180
181 AIRGWPPSKA SDAQQLGTRL CNLDLSPENE LILVDLTEDG FMDTEQDEVT QRVDKDFTIF 240
241 IQFESDPPLT LSLPGRMTVQ EVKMNVFDIK SIPVRHQEWT GWPNGCDNDT TLAQSGIGLS 300
301 HRFAVRSTER SAPTNSNQHN AFDVVTVDSE SSTDEFEDAT DFNNAEYIFT DSPPAQPPNR 360
361 HLIPNDTDDE ISGSTQFVEN YKARYGEPCP EFFVGSLENA KQLACLRSAK ERKLLAIYLH 420
421 HGKSILSNVF CDQLMKHESI IQTFKEKFVL YGWDMTYESN KDMFLSSLTA CISSNASLTA 480
481 RNIKLDKLPA IMLVGKSRQL GSNCEVLSVI HGNIGLDDLL TRLIETCEMF EEQLQVEIRQ 540
541 EDERAARDQV KAEQDMAYQE TLQADMAKDA AKRQKEAAQL AERKRMESER AEEDARRESI 600
601 RLVAQQSLPQ EPSEQETGTS KIRVRKPTGD FLERRFFINN NLQDLLNFVT ANGFLIEEYK 660
661 LISSWPRRDL TAIESSQTLE SLKLYPQETV ILEER |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
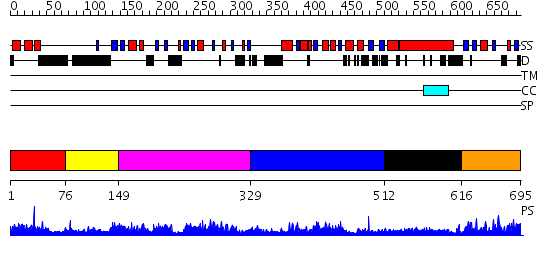
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..75] | 1.55 | No description for 2damA was found. | |
| 2 | View Details | [76..148] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [149..328] | 19.154902 | Crystal Structure of ISG15, the Interferon-Induced Ubiquitin Cross Reactive Protein | |
| 4 | View Details | [329..511] | 23.522879 | No description for 2dlxA was found. | |
| 5 | View Details | [512..615] | 6.39794 | No description for 2i1jA was found. | |
| 6 | View Details | [616..695] | 21.69897 | Fas-assosiated factor 1, Faf1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)