













| General Information: |
|
| Name(s) found: |
LDHB
[HGNC (HUGO)]
|
| Description(s) found:
Found 43 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 334 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATLKEKLIA PVAEEEATVP NNKITVVGVG QVGMACAISI LGKSLADELA LVDVLEDKLK 60
61 GEMMDLQHGS LFLQTPKIVA DKDYSVTANS KIVVVTAGVR QQEGESRLNL VQRNVNVFKF 120
121 IIPQIVKYSP DCIIIVVSNP VDILTYVTWK LSGLPKHRVI GSGCNLDSAR FRYLMAEKLG 180
181 IHPSSCHGWI LGEHGDSSVA VWSGVNVAGV SLQELNPEMG TDNDSENWKE VHKMVVESAY 240
241 EVIKLKGYTN WAIGLSVADL IESMLKNLSR IHPVSTMVKG MYGIENEVFL SLPCILNARG 300
301 LTSVINQKLK DDEVAQLKKS ADTLWDIQKD LKDL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
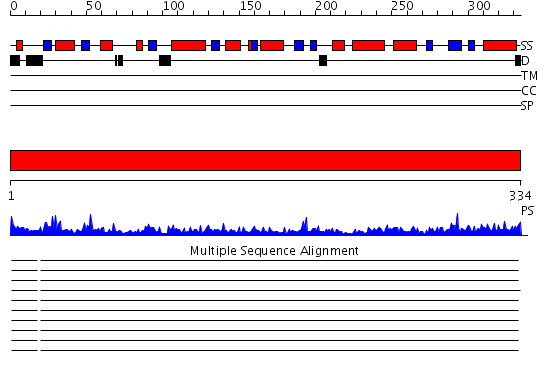
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..334] | 104.0 | Human B lactate dehydrogenase complexed with NAD+ and 4-hydroxy-1,2,5-oxadiazole-3-carboxylic acid |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)