













| General Information: |
|
| Name(s) found: |
ACTB
[HGNC (HUGO)]
|
| Description(s) found:
Found 46 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 375 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDDDIAALVV DNGSGMCKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS 60
61 KRGILTLKYP IEHGIVTNWD DMEKIWHHTF YNELRVAPEE HPVLLTEAPL NPKANREKMT 120
121 QIMFETFNTP AMYVAIQAVL SLYASGRTTG IVMDSGDGVT HTVPIYEGYA LPHAILRLDL 180
181 AGRDLTDYLM KILTERGYSF TTTAEREIVR DIKEKLCYVA LDFEQEMATA ASSSSLEKSY 240
241 ELPDGQVITI GNERFRCPEA LFQPSFLGME SCGIHETTFN SIMKCDVDIR KDLYANTVLS 300
301 GGTTMYPGIA DRMQKEITAL APSTMKIKII APPERKYSVW IGGSILASLS TFQQMWISKQ 360
361 EYDESGPSIV HRKCF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | No Comments | Foltz DR, et al (2006) | |
| View Run | Sample pIC145 from april 2005 | Okada, M., et al. (2006) | |
| View Run | Sample pIC146 from april 2005 | Okada, M., et al. (2006) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
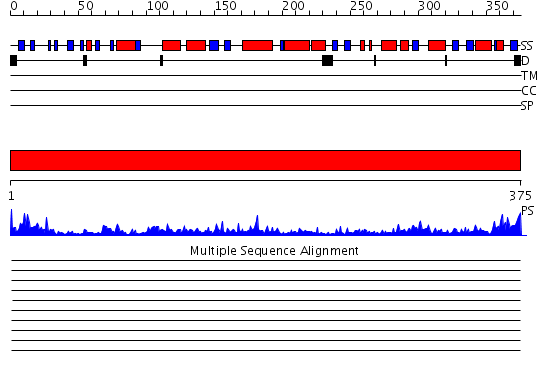
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..375] | 146.0 | No description for 2oanA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.76 |
Source: Reynolds et al. (2008)