













| General Information: |
|
| Name(s) found: |
GIP3 /
YPL137C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1276 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA endoplasmic reticulum [IDA ribosome [IDA |
| Biological Process: |
chromosome segregation
[IMP regulation of phosphoprotein phosphatase activity [IMP |
| Molecular Function: |
protein phosphatase type 1 regulator activity
[IMP protein phosphatase 1 binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MITNTEFDVP VDWLYKGKSR RKTNTKPSRP STSPASSSST SSSKNGDNST SGNRSSNDKP 60
61 RARSSSVSNA ALCNTEKPDL KRNDGNTSAS DTDNIPLLTP INSGNRSDSA DIDNPATVDA 120
121 IDLIDNDDNG SSTQFVRKKR STSISNAVVS SKPRLASSAI NATASSSVGK GKHPPISSPS 180
181 NATLKRSNST SGEKTKRSIF GSLFSKRSTS SSASTAKKPL PVVNTSTTEN ESGGIKAVAT 240
241 PDPRVKEISS PMRGVAPTAS KPQTPILPSP ALAVKDLSTV SLKRVSFAVD KFESDPPQQL 300
301 PSRTPKKGNI LIPDDMISEV PSISVGISSS NQSAKSTNSN IKGPLYTKKS KEYILALENQ 360
361 KLALREAAKH QQEAHFAANR IAFEVANFKT ASDAGGKLTE KSSEGTITKQ REEVSPPNVE 420
421 ADRELENNKL AENLSKAGID KPIHMHEHYF KEPDQDKYQD GHSIENNEVT LDVIYTRCCH 480
481 LREILPIPST LRQVKDKTAP LQILKFLNPK PTLIDILSFC DFITIAPIHT IVFDNVALNQ 540
541 DMFRIIISAL VNSTVLDKLS LRNVRIDQDG WKLLCKFLLL NKSLNKLDIS QTKIKSDLAE 600
601 SLYRHNMDWN LFTDVLSQRS HKPIEELLFN GIQFSKIPYS CFARLLTSFA TQKNFPESGI 660
661 RLGLAGATTS NISQDCLKFI FNWMSQYNVQ GVDLAFNDLS TMIKPMVGKL SALSYDNLRY 720
721 FILNSTNIST SYDLALLLKY LSKLPNLIFL DLSNLSQCFP DILPYMYKYL PRFPNLKRIH 780
781 LDSNNLTLKE LAVVCNILIK CKSLSHVSMT NQNVENFYLM NGTDSPVQQT NTDGDLDSSS 840
841 TLDVKGQFAK NSFSSTLYAF ARDSPNLIGL DFDYDLISEE IQSRIALCLM RNMKRTMDST 900
901 FQLDELDSQD DLLFDGSLVT MTAESVLEKL NLLSDKSTKV KKDTTKRYLL KKYIEKFHIL 960
961 HHNVQHTIDT MFEKRKSGEL PLQEKENLVR LLLLEQNLCN ILELFSHNPN LNDVLGSSRD1020
1021 DSKESVDSSE DSKLPALKHV ESGYHVPEEK IQPENDVITA RPHLMATDSG KTIDVFTGKP1080
1081 LVFKHTSSST SVGCKKQEEE EGELHKWGFF VQQQRSLYPE NESTRQTPFA SGDTPINTET1140
1141 AGKSTSSPSV STSNNETATT SLFSPANPKI LPKIPSGAVL RSAIMKAKGI DSIDDLIQNV1200
1201 NSNNIELENI YGESIQNSAS TFTPGVDSDV SAPNTDKGSV ETLPAVSTDD PNCEVKVTAT1260
1261 YDKLLNNLSM ERSIRL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
BNI4 BUD14 GIP3 GLC7 GLC8 HER1 PPZ1 PUF3 SDS22 SOL1 SOL2 YLR257W YPI1 |
| View Details | Qiu et al. (2008) |

|
GIP1 GIP3 GIP4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
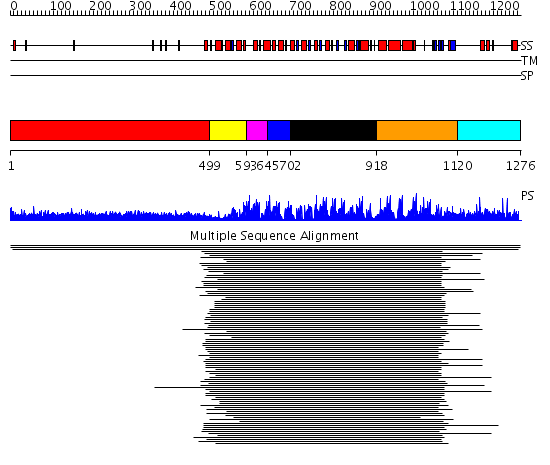
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..498] | 94.901997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [499..592] | 21.0 | Ribonuclease inhibitor | |
| 3 | View Details | [593..644] | 21.0 | Ribonuclease inhibitor | |
| 4 | View Details | [645..701] | 21.0 | Ribonuclease inhibitor | |
| 5 | View Details | [702..917] | 21.0 | Ribonuclease inhibitor | |
| 6 | View Details | [918..1119] | 19.923995 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1120..1276] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)