













| General Information: |
|
| Name(s) found: |
SFL1 /
YOR140W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 766 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear chromosome
[IPI |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[IMP |
| Molecular Function: |
specific transcriptional repressor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEEETVSAP APASTPAPAG TDVGSGGAAA GIANAGAEGG DGAEDVKKHG SKMLVGPRPP 60
61 QNAIFIHKLY QILEDESLHD LIWWTPSGLS FMIKPVERFS KALATYFKHT NITSFVRQLN 120
121 IYGFHKVSHD HSSNDANSGD DANTNDDSNT HDDNSGNKNS SGDENTGGGV QEKEKSNPTK 180
181 IWEFKHSSGI FKKGDIEGLK HIKRRASSRN NSSINSRKNS SNQNYDIDSG ARVRPSSIQD 240
241 PSTSSNSFGN FVPQIPGANN SIPEYFNNSH VTYENANHAP LESNNPEMQE QNRPPNFQDE 300
301 TLKHLKEINF DMVKIIESMQ HFISLQHSFC SQSFTFKNVS KKKSENIVKD HQKQLQAFES 360
361 DMLTFKQHVM SRAHRTIDSL CAVNAAATAA SVAPAPAPTS TSAYAPKSQY EMMVPPGNQY 420
421 VPQKSSSTTN IPSRFNTASV PPSQLLYNTN RSRNQHVTYA SEPAHVPNFI NQPIPIQQLP 480
481 PQYADTFSTP QMMHNPFASK NNNKPGNTKR TNSVLMDPLT PAASVGVQGP LNYPIMNINP 540
541 SVRDYNKPVP QNMAPSPIYP INEPTTRLYS QPKMRSLGST SSLPNDRRNS PLKLTPRSSL 600
601 NEDSLYPKPR NSLKSSISGT SLSSSFTLVA NNPAPIRYSQ QGLLRSLNKA ANCAPDSVTP 660
661 LDSSVLTGPP PKNMDNLPAV SSNLINSPMN VEHSSSLSQA EPAPQIELPQ PSLPTTSTTK 720
721 NTGEADNSKR KGSGVYSLLN QEDSSTSSAD PKTEDKAAPA LKKVKM |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
BIM1 BMH1 GTS1 HAP1 RCN2 SFL1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
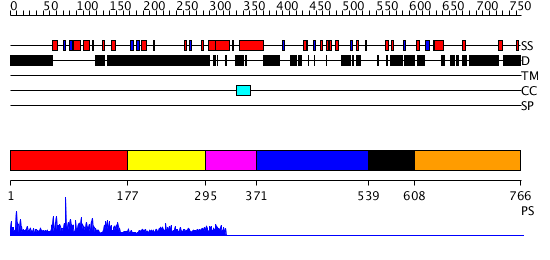
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..176] | 144.325697 | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | |
| 2 | View Details | [177..294] | 1.077987 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [295..370] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [371..538] | 14.81 | No description for 1ldvA was found. | |
| 5 | View Details | [539..607] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [608..766] | 11.0 | No description for 1deqA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)