













| General Information: |
|
| Name(s) found: |
PDR16 /
YNL231C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 351 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA cytoplasm [IDA microsome [IDA |
| Biological Process: |
response to drug
[IGI phospholipid transport [IDA sterol biosynthetic process [IMP |
| Molecular Function: |
phosphatidylinositol transporter activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFKRFSKKKE APEDPKNLIN IDKPIKELPA SIAIPKEKPL TGEQQKMYDE VLKHFSNPDL 60
61 KVYTSEKNKS EDDLKPLEEE EKAWLTRECF LRYLRATKWV LKDCIDRITM TLAWRREFGI 120
121 SHLGEEHGDK ITADLVAVEN ESGKQVILGY ENDARPILYL KPGRQNTKTS HRQVQHLVFM 180
181 LERVIDFMPA GQDSLALLID FKDYPDVPKV PGNSKIPPIG VGKEVLHILQ THYPERLGKA 240
241 LLTNIPWLAW TFLKLIHPFI DPLTREKLVF DEPFVKYVPK NELDSLYGGD LKFKYNHDVY 300
301 WPALVETARE KRDHYFKRFQ SFGGIVGLSE VDLRGTHEKL LYPVKSESST V |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
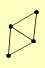
|
CDC50 FAT1 PDR16 SFH5 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |

Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..57] [113..351] |
139.68867 | N-terminal domain of phosphatidylinositol transfer protein sec14p; C-terminal domain of phosphatidylinositol transfer protein sec14p | |
| 2 | View Details | [58..112] | 139.68867 | N-terminal domain of phosphatidylinositol transfer protein sec14p; C-terminal domain of phosphatidylinositol transfer protein sec14p |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)