













| General Information: |
|
| Name(s) found: |
PDR8 /
YLR266C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 701 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IPI |
| Biological Process: |
positive regulation of transcription from RNA polymerase II promoter
[IGI response to stress [IGI |
| Molecular Function: |
DNA binding
[IDA specific RNA polymerase II transcription factor activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDGSHFPMKS TTGEPVSSGK KGKRRKVIKS CAFCRKRKLK CSQARPMCQQ CVIRKLPQCV 60
61 YTEEFNYPLS NTELFEQVPN VALVQKIENL QTLLKENDNN NAKPVYCRSS ENPLRSLRTS 120
121 VLGDNGSRYV FGPTSWKTLS LFEQNKFQTE FQNLWKVLKP LPECTKSQLN ENDVVADLPS 180
181 FPQMESCIKS FFAGPLFGIL HIFNQDDILS LLDRLFIRDT TDKNLVILLD LQGNAKDKYN 240
241 LGIVLQILCL GYYNQDIPSS VSHFLHTLSA ASLSSSSSNF VEKLQFFLLS YISVMINCTD 300
301 GVWDATQGVD LINELCQGCI SLGLNDIDKW YLNESEETKQ NLRCIWFWAL FLDVSTSYDI 360
361 GNPPSISDDL LDLSIFTAQN FQSPSIDFRR VKLMHDFLDV SRFTTREIHK REMNEKLTTF 420
421 SLRLIEFIQS NFSPIEHYTN SVYYSDIDPF DILILSRSLS IVASIYNIEM IIAQQSRIID 480
481 KNRMVQFLLI SISVCVNTMV FHFKEPINDQ ENVLTEGLKL SIILINPLLI RIVSQVYSLA 540
541 FNRLIFREKA FLFLIDLDTG KKIQFIKYEE ENFDELLTGF DVRTDKFLSF SGTIIRFYEI 600
601 IDSIFAVNER NKRLLKAVSN FYQLTSTLAF ERVSRVLFDK ASQARIETEK IWLKKGINME 660
661 HFSDLMIEDF INDVWKTFKE ISKDLWSIDK KKFYKQYHFD L |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
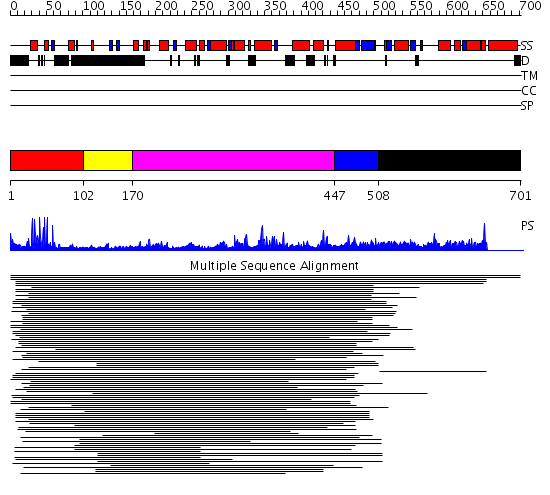
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | 68.67837 | Hap1 (Cyp1); HAP1 | |
| 2 | View Details | [102..169] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [170..446] | 6.120992 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [447..507] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [508..701] | 1.026958 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.65 |
Source: Reynolds et al. (2008)