













| General Information: |
|
| Name(s) found: |
ALY1 /
YKR021W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 915 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLQFNTENDT VAPVFPMEQD INAAPDAVPL VQTTTLQVFV KLAEPIVFLK GFETNGLSEI 60
61 APSILRGSLI VRVLKPNKLK SISITFKGIS RTEWPEGIPP KREEFSDVET VVNHTWPFYQ 120
121 ADDGMNSFTL EHHSSNNSSN RPSMSDEDYL LEKSGASVYI PPTAEPPKDN SNLSLDAYER 180
181 NSLSSDNLSN KPVSSDVSHD DSKLLAIQKT PLPSSSRRGS VPANFHGNSL SPHTFISDLF 240
241 TKTFSNSGAT PSPEQEDNYL TPSKDSKEVF IFRPGDYIYT FEQPISQSYP ESIKANFGSV 300
301 EYKLSIDIER FGAFKSTIHT QLPIKVVRLP SDGSVEETEA IAISKDWKDL LHYDVVIFSK 360
361 EIVLNAFLPI DFHFAPLDKV TLHRIRIYLT ESMEYTCNSN GNHEKARRLE PTKKFLLAEH 420
421 NGPKLPHIPA GSNPLKAKNR GNILLDEKSG DLVNKDFQFE VFVPSKFTNS IRLHPDTNYD 480
481 KIKAHHWIKI CLRLSKKYGD NRKHFEISID SPIHILNQLC SHANTLLPSY ESHFQYCDED 540
541 GNFAPAADQQ NYASHHDSNI FFPKEVLSSP VLSPNVQKMN IRIPSDLPVV RNRAESVKKS 600
601 KSDNTSKKND QSSNVFASKQ LVANIYKPNQ IPRELTSPQA LPLSPITSPI LNYQPLSNSP 660
661 PPDFDFDLAK RGAADSHAIP VDPPSYFDVL KADGIELPYY DTSSSKIPEL KLNKSRETLA 720
721 SIEEDSFNGW SQIDDLSDED DNDGDIASGF NFKLSTSAPS ENVNSHTPIL QSLNMSLDGR 780
781 KKNRASLHAT SVLPSTIRQN NQHFNDINQM LGSSDEDAFP KSQSLNFNKK LPILKINDNV 840
841 IQSNSNSNNR VDNPEDTVDS SVDITAFYDP RMSSDSKFDW EVSKNHVDPA AYSVNVASEN 900
901 RVLDDFKKAF REKRK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
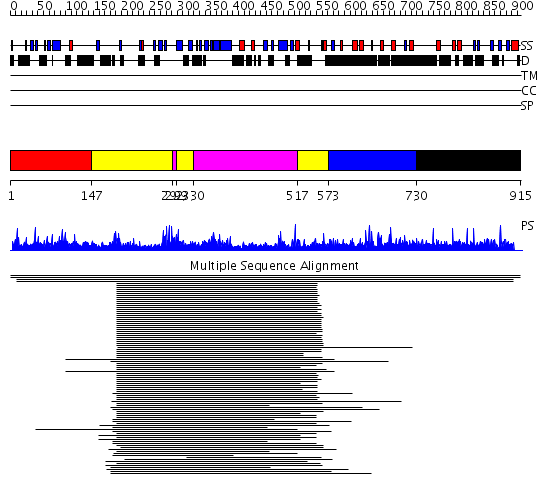
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..146] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [147..291] [299..329] [517..572] |
87.522879 | Arrestin | |
| 3 | View Details | [292..298] [330..516] |
87.522879 | Arrestin | |
| 4 | View Details | [573..729] | 3.038997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [730..915] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)