













| General Information: |
|
| Name(s) found: |
APN1 /
YKL114C
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 367 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrion [IDA |
| Biological Process: |
base-excision repair
[IDA telomere maintenance [IMP |
| Molecular Function: |
DNA-(apurinic or apyrimidinic site) lyase activity
[IDA double-stranded DNA specific 3'-5' exodeoxyribonuclease activity [IDA phosphoric diester hydrolase activity [IDA 3'-tyrosyl-DNA phosphodiesterase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPSTPSFVRS AVSKYKFGAH MSGAGGISNS VTNAFNTGCN SFAMFLKSPR KWVSPQYTQE 60
61 EIDKFKKNCA TYNYNPLTDV LPHGQYFINL ANPDREKAEK SYESFMDDLN RCEQLGIGLY 120
121 NLHPGSTLKG DHQLQLKQLA SYLNKAIKET KFVKIVLENM AGTGNLVGSS LVDLKEVIGM 180
181 IEDKSRIGVC IDTCHTFAAG YDISTTETFN NFWKEFNDVI GFKYLSAVHL NDSKAPLGAN 240
241 RDLHERLGQG YLGIDVFRMI AHSEYLQGIP IVLETPYEND EGYGNEIKLM EWLESKSESE 300
301 LLEDKEYKEK NDTLQKLGAK SRKEQLDKFE VKQKKRAGGT KRKKATAEPS DNDILSQMTK 360
361 KRKTKKE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
APN1 YPD1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
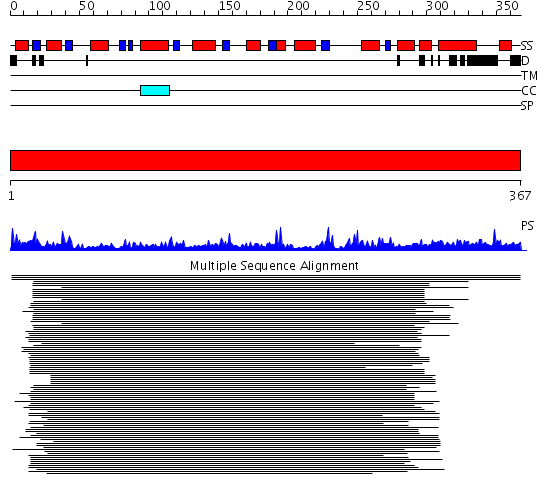
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..367] | 688.67837 | Endonuclease IV |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)