













| General Information: |
|
| Name(s) found: |
HIR3 /
YJR140C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1648 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA HIR complex [IDA |
| Biological Process: |
regulation of transcription involved in G1/S-phase of mitotic cell cycle
[IMP DNA replication-independent nucleosome assembly [IDA RNA elongation from RNA polymerase II promoter [IGI |
| Molecular Function: |
transcription corepressor activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSMFNALNSN IEGEQYEAEE HSRELQIEQS FNILQDALID LKNKDFEKSD SKFQELFQID 60
61 VVKPDRWGMY RNSSPTLDNL RYLCYRNRGM YYHLYLENNY ERLNSQELVN CILKAVENLV 120
121 ESIQHSDADF AVTDLLARIF KSFNSVKLER LISEYEFTKQ ENLSLLLGRH RKFLLNDLTL 180
181 MMNNYVELTN KLLVPNLSDN TIFERYHLEK YKDIKPEPLA FGPILSRISE MKKQDEEIMK 240
241 KLDVFNVTLN EESWDEVAKA LKNLLPSVKT SSLIGRNMDP YNEIEEPIEA VKFELSEAIN 300
301 NTPSLDRESE RQEEEQDNES VRADDKSGNL APSDIQTNEE ARPNKRTDEH IDSTKPLQRS 360
361 SKRFKEREQE NSKELVMDVH KRFFGEFNTL LSYIHILPFC DFDTFASKFI IGSSDKQPEK 420
421 FIPYTDLYEC LKSWSSRYTD IFNQNDYLSS GSNENEELFQ LNALLKSNAF DDKESFPRYL 480
481 NDLDSDHIRS FISEVNAGNL HFHQVRLKLL FKLLGTYDEG NGRRLIIDYL WESQLLKIVL 540
541 WFVFGIESNI FALINKNKRQ CKYLALSIYE LLVNHLGNIV EEITNKRIQG HKSADLKSQR 600
601 NKVEKRIRSW HTLLEQIADE KDKELYVHFQ WTHYCFLQYT CDIVDSRLSE TLTSLENTIK 660
661 DSDSSLDIAY PNYRHIPALN LNTVQSQKRK IRIIQNITVE DISEDTNSDT HSENHLETLE 720
721 KVLLHILHPS TNHSNIDEEM VSFIFNSPFL LKIRLWGVLF SSYVKKSSIQ DVQRIYFHVL 780
781 DFMKGALTSP VYKESNPHGR HQMLLTVLTA IGYLSSQLTA ILNSNRWESS DFVLEDYMFE 840
841 KLLQTFFFFY TVLFYESSAV NDVSNKSFFK RASKSSGKMK DIMIDLATLI LYYYDLQAKL 900
901 RTPAEQGIET TELIWSLHTL FGHFHFCDAS NGKFLDLAEK LLCQFINNDS FLQLKQILWC 960
961 RYHYAIASDN FSPDLHDTKA VEMEKIHSLP LGTYLIKLQY QNKNPYLSSS KTTLKQIMDN1020
1021 IIEKIGDPST LDNHIISRNS FLLNEYLSRP ITADLLKHTF SGATSLYLTS PNDELQQGMT1080
1081 AGLFYVSSLQ SLGLYKMRKK SMQARPSELD SIIRMLKNDI IYNTNRFESW ILLGKCYSYI1140
1141 VEDDLIWTSD KITVPEKKDV IALTQRKAIL CYLMAISIYY SKLDRTIDDK KIILEALDDL1200
1201 GSMLISGYYN PMNKLCFSWK SSAENTMRLS ETGEVVMEKT KKITTISDFN IEQSIFLCFN1260
1261 RACSLSGDIK SQDDVFVLNW SSFYNLAKFF FKTDGGNNCK LVAKYITQGC QIAYESSPAK1320
1321 DPIIEPHYLL VNACYKWVKR GVIGVNEALT LLSKDNQFFQ EQEEFWVNDE GLAWDYQEKF1380
1381 FFDKIIRLLR HLLSVDKKKW QHRPRYRIAR ILFDDLGDVN GALEEMDSLI SAKSINKNLV1440
1441 NIWKPDFERP GKHFIYTYQY LVLYLDLLFA IKDFNTTGLV IKKLRRFGSG TVNVNELLER1500
1501 AINVYTQSAK IKLQLQDKSY VEQILPTLNY QEFLKISEQL NQVFDQGKYP EEISSGLKLA1560
1561 FQLKKGHSGI AFDSVCLGIY FEYLYFPLAR QDQSLTDVND ENNPALPSSG SVTSKSTPDP1620
1621 TSKPSAIKKR VTKKEVFDRV RLLVDKIT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
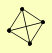
|
ASF1 HIR2 HIR3 RAD53 |
| View Details | Krogan NJ, et al. (2006) |
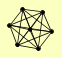
|
ASF1 HIR1 HIR2 HIR3 HPC2 QDR2 SFP1 |
| View Details | Gavin AC, et al. (2006) |

|
HIR3 MAM33 RAD53 |
| View Details | Gavin AC, et al. (2002) |
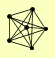
|
ASF1 HIR3 MAM33 RAD53 RNR2 RNR4 |
| View Details | Ho Y, et al. (2002) |

|
ADH3 CDC10 ECM10 HIR3 MGM101 PYC1 RFA2 SML1 |
| View Details | Qiu et al. (2008) |

|
ASF1 CAC2 HIR1 HIR2 HIR3 HPC2 MSI1 RLF2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Green EM, et al (2005) | |
| View Run | sample 3121 run on DecaLCQ: searched for phosphorylation | Green EM, et al (2005) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |

Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..521] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [522..1648] | 1.003001 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [498..559] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [560..733] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [734..909] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [910..1027] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1028..1247] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1248..1355] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1356..1648] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)