













| General Information: |
|
| Name(s) found: |
RPH1 /
YER169W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 796 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[IDA DNA repair [IMP |
| Molecular Function: |
specific transcriptional repressor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTKLIAPSEI VGGVPVFKPT YEQFEDFYAY CKAINKYGMK SGVVKVIPPK EWKDKLDLPY 60
61 SAETLQKIKI KSPIQQHISG NKGLFMVQNV EKNKTYNIIQ WKDLSKDYVP PEDPKARRNS 120
121 RKGSVSKSTK LKLKNFESSF NIDDFEQFRT EYTIDLSDFQ NTERLKFLEE YYWKTLNFTT 180
181 PMYGADTPGS IFPEGLNVWN VAKLPNILDH METKVPGVND SYLYAGLWKA SFSWHLEDQD 240
241 LYSINYIHFG APKQWYSIPQ EDRFKFYKFM QEQFPEEAKN CPEFLRHKMF LASPKLLQEN 300
301 GIRCNEIVHH EGEFMITYPY GYHAGFNYGY NLAESVNFAL EEWLPIGKKA GKCHCISDSV 360
361 EIDVKKLAKS WRDNNKESKG TPPLNQLPNP AMPLLHRPTL KEMESSSLRS TSPDVGHFSN 420
421 FKSKSSGVSS PLLSRMKDYS NIVEPTLEDP TLKLKRISSF QEQPLNKLLK RETSQTAMLT 480
481 DHEDNIVAMS LTSMANSAAS SPRLPLSRLN SSNELSNAQP LLDMTNNTLA FPRPNGPSGL 540
541 NPLLYISNKN ISGISHSAPH SPVNPNISLI KRVKSPNIVT LNISRESSRS PIALNYEARQ 600
601 QHSQQHSFST PSTVSNLSTS VLGPLSDTND IKTPHPERPN HKTANRILKK ESPVETSKSN 660
661 LILSKVASTR QEDSFTSRND DLDKEQGSSP LNSKFAPEEI VLSGKNKIYI CKECQRKFSS 720
721 GHHLTRHKKS VHSGEKPHSC PKCGKRFKRR DHVLQHLNKK IPCISNETTV DAPIMNPTVQ 780
781 PQDGKAAINQ QSTPLN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
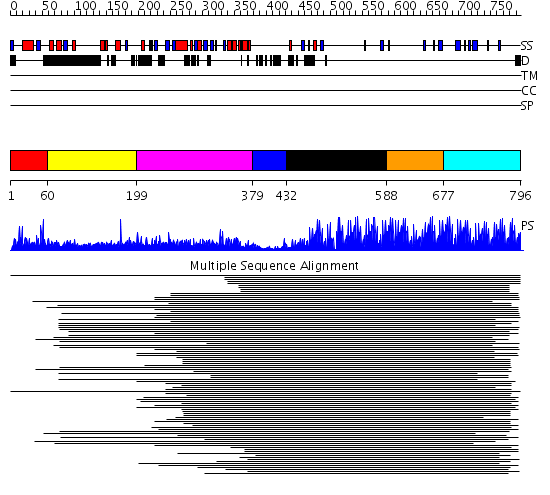
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..59] | 24.920819 | jmjN domain No confident structure predictions are available. | |
| 2 | View Details | [60..198] | 3.084997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [199..378] | 57.05061 | JmjC domain No confident structure predictions are available. | |
| 4 | View Details | [379..431] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [432..587] | 55.927757 | Ying-yang 1 (yy1, zinc finger domain) | |
| 6 | View Details | [588..676] | 8.174996 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [677..796] | 94.9897 | Designed zinc finger protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)