













| General Information: |
|
| Name(s) found: |
ISC1 /
YER019W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 477 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA endoplasmic reticulum [IDA |
| Biological Process: |
response to salt stress
[IMP sphingolipid catabolic process [IDA |
| Molecular Function: |
phospholipase C activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYNRKDRDVH ERKEDGQSEF EALNGTNAIM SDNSKAYSIK FLTFNTWGLK YVSKHRKERL 60
61 RAIADKLAGH SMLTPISDEL LPNGGDSNEN EDYDVIALQE IWCVEDWKYL ASACASKYPY 120
121 QRLFHSGILT GPGLAILSKV PIESTFLYRF PINGRPSAVF RGDWYVGKSI AITVLNTGTR 180
181 PIAIMNSHMH APYAKQGDAA YLCHRSCQAW DFSRLIKLYR QAGYAVIVVG DLNSRPGSLP 240
241 HKFLTQEAGL VDSWEQLHGK QDLAVIARLS PLQQLLKGCT TCDSLLNTWR AQRQPDEACR 300
301 LDYALIDPDF LQTVDAGVRF TERIPHLDCS VSDHFAYSCT LNIVPQGTES RPSTSVKRAK 360
361 THDRELILQR YSNYETMIEC IHTYLKTAQR QKFFRGLHFW ASILLLIASL VVTTFTANKA 420
421 GWSSIFWVLF AIAVSISGTI DGAISFLFGR SEIRALIEVE QEVLDAEHHL QTFLSEK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
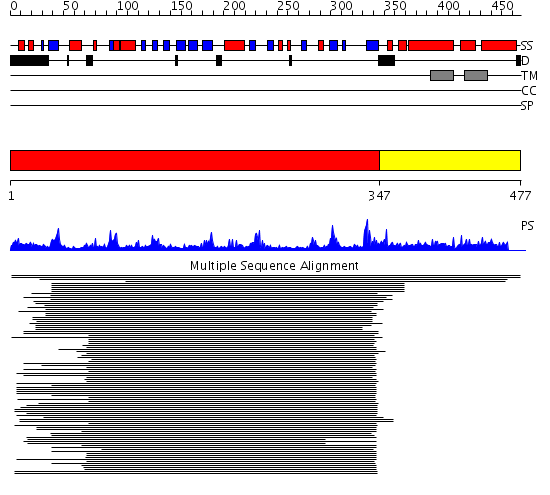
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..346] | 41.39794 | DNA repair endonuclease Hap1 | |
| 2 | View Details | [347..477] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|