













| General Information: |
|
| Name(s) found: |
GLC3 /
YEL011W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 704 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
glycogen biosynthetic process
[IMP |
| Molecular Function: |
1,4-alpha-glucan branching enzyme activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYNIPDNVKG AVEFDPWLKP FADVLSERRY LADKWLYDIT HATPDGSYQS LSKFARDSYK 60
61 SYGLHANPET KEITYKEWAP NAERAFLVGD FNNWDTTSHE LKNKDEFGNF TITLHPLPNG 120
121 DFAIPHDSKI KVMFILPDGS KIFRLPAWIT RATQPSKETS KQFGPAYEGR FWNPENPYKF 180
181 VHPRPKFSES VDSLRIYEAH VGISSPEPKI TTYKEFTEKV LPRIKYLGYD AIQLMAIMEH 240
241 AYYASFGYQV TNFFAASSRF GTPEELKELI DTAHSMGILV LLDVVHSHAS KNVEDGLNMF 300
301 DGSDHQYFHS ISSGRGEHPL WDSRLFNYGK FEVQRFLLAN LAFYVDVYQF DGFRFDGVTS 360
361 MLYVHHGVGA GGSFSGDYNE YLSRDRSFVD HEALAYLMLA NDLVHEMLPN LAVTVAEDVS 420
421 GYPTLCLPRS IGGTGFDYRL AMALPDMWIK LIKEKKDDEW EMGSIVYTLT NRRYGEKVVA 480
481 YCESHDQALV GDKTLAFWLM DAAMYTDMTV LKEPSIVIDR GIALHKMIRL ITHSLGGEAY 540
541 LNFEGNEFGH PEWLDFPNVN NGDSYKYARR QFNLADDPLL RYQNLNEFDR SMQLCEKRHK 600
601 WLNTKQAYVS LKHEGDKMIV FERNNLLFIF NFHPTNSYSD YRVGVEKAGT YHIVLNSDRA 660
661 EFGGHNRINE SSEFFTTDLE WNNRKNFLQV YIPSRVALVL ALKE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
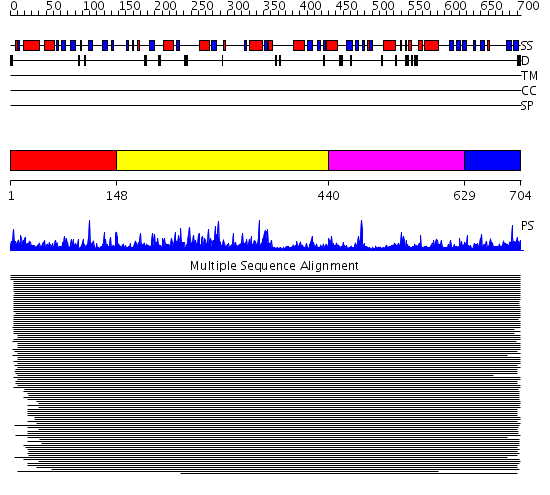
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | 206.522879 | Glycosyltrehalose trehalohydrolase, N-terminal domain N; Glycosyltrehalose trehalohydrolase; Glycosyltrehalose trehalohydrolase, central domain | |
| 2 | View Details | [148..439] | 206.522879 | Glycosyltrehalose trehalohydrolase, N-terminal domain N; Glycosyltrehalose trehalohydrolase; Glycosyltrehalose trehalohydrolase, central domain | |
| 3 | View Details | [440..628] | 206.522879 | Glycosyltrehalose trehalohydrolase, N-terminal domain N; Glycosyltrehalose trehalohydrolase; Glycosyltrehalose trehalohydrolase, central domain | |
| 4 | View Details | [629..704] | 100.751666 | Cyclomaltodextrin glycanotransferase, domain D; Cyclodextrin glycosyltransferase, C-terminal domain; Cyclodextrin glycosyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)