













| General Information: |
|
| Name(s) found: |
HST4 /
YDR191W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 370 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
histone deacetylation
[IMP chromatin silencing at telomere [IGI short-chain fatty acid metabolic process [IMP |
| Molecular Function: |
DNA binding
[ISS histone deacetylase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKQKFVLPIT PPSTAEKKPQ TENRCNENLK PRRLLPQLKK SVRNRKPRLS YRPELNSVFD 60
61 LDAYVDSTHL SKSQRHHMDR DAGFISYALN YSKRMVVVSG AGISVAAGIP DFRSSEGIFS 120
121 TVNGGSGKDL FDYNRVYGDE SMSLKFNQLM VSLFRLSKNC QPTKFHEMLN EFARDGRLLR 180
181 LYTQNIDGLD TQLPHLSTNV PLAKPIPSTV QLHGSIKHME CNKCLNIKPF DPELFKCDDK 240
241 FDSRTEIIPS CPQCEEYETV RKMAGLRSTG VGKLRPRVIL YNEVHPEGDF IGEIANNDLK 300
301 KRIDCLIIVG TSLKIPGVKN ICRQFAAKVH ANRGIVLYLN TSMPPKNVLD SLKFVDLVVL 360
361 GDCQHVTSLL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
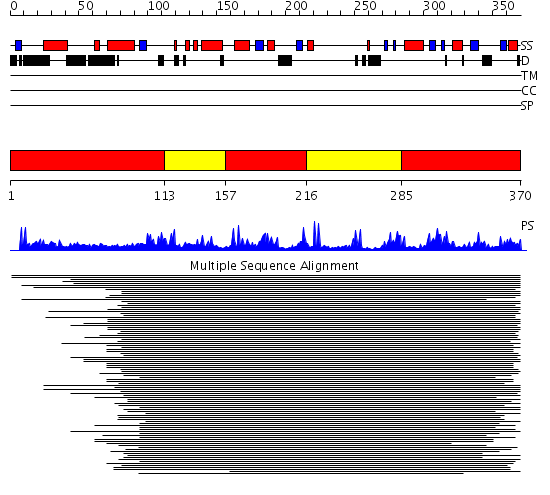
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..112] [157..215] [285..370] |
143.035458 | AF1676, Sir2 homolog (Sir2-AF1?) | |
| 2 | View Details | [113..156] [216..284] |
143.035458 | AF1676, Sir2 homolog (Sir2-AF1?) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.59 |
Source: Reynolds et al. (2008)