













| General Information: |
|
| Name(s) found: |
IDP1 /
YDL066W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 428 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial nucleoid [IDA |
| Biological Process: |
glutamate biosynthetic process
[IGI isocitrate metabolic process [TAS |
| Molecular Function: |
isocitrate dehydrogenase (NADP+) activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSMLSRRLFS TSRLAAFSKI KVKQPVVELD GDEMTRIIWD KIKKKLILPY LDVDLKYYDL 60
61 SVESRDATSD KITQDAAEAI KKYGVGIKCA TITPDEARVK EFNLHKMWKS PNGTIRNILG 120
121 GTVFREPIVI PRIPRLVPRW EKPIIIGRHA HGDQYKATDT LIPGPGSLEL VYKPSDPTTA 180
181 QPQTLKVYDY KGSGVAMAMY NTDESIEGFA HSSFKLAIDK KLNLFLSTKN TILKKYDGRF 240
241 KDIFQEVYEA QYKSKFEQLG IHYEHRLIDD MVAQMIKSKG GFIMALKNYD GDVQSDIVAQ 300
301 GFGSLGLMTS ILVTPDGKTF ESEAAHGTVT RHYRKYQKGE ETSTNSIASI FAWSRGLLKR 360
361 GELDNTPALC KFANILESAT LNTVQQDGIM TKDLALACGN NERSAYVTTE EFLDAVEKRL 420
421 QKEIKSIE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
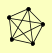
|
DDR48 FCP1 IDP1 YMR118C ZEO1 |
| View Details | Qiu et al. (2008) |
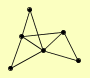
|
ARG5,6 IDH1 IDH2 IDP1 ILV5 KGD1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
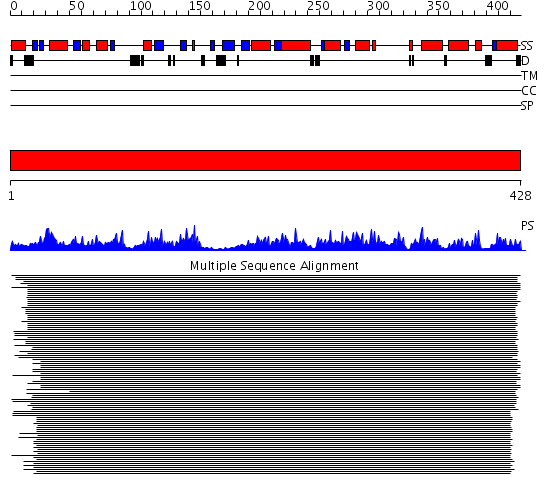
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..428] | 158.228787 | 3-isopropylmalate dehydrogenase, IPMDH |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)