













| General Information: |
|
| Name(s) found: |
FES1 /
YBR101C
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 290 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
translation
[IMP |
| Molecular Function: |
adenyl-nucleotide exchange factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKLLQWSIA NSQGDKEAMA RAGQPDPKLL QQLFGGGGPD DPTLMKESMA VIMNPEVDLE 60
61 TKLVAFDNFE MLIENLDNAN NIENLKLWEP LLDVLVQTKD EELRAAALSI IGTAVQNNLD 120
121 SQNNFMKYDN GLRSLIEIAS DKTKPLDVRT KAFYALSNLI RNHKDISEKF FKLNGLDCIA 180
181 PVLSDNTAKP KLKMRAIALL TAYLSSVKID ENIISVLRKD GVIESTIECL SDESNLNIID 240
241 RVLSFLSHLI SSGIKFNEQE LHKLNEGYKH IEPLKDRLNE DDYLAVKYVL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
FES1 LSG1 LYS2 |
| View Details | Gavin AC, et al. (2002) |
|
FES1 HSP42 TIF1, TIF2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
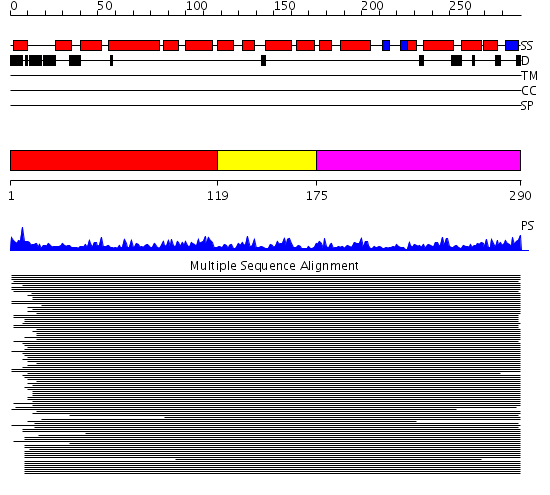
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] | 56.221849 | Importin alpha | |
| 2 | View Details | [119..174] | 56.221849 | Importin alpha | |
| 3 | View Details | [175..290] | 56.221849 | Importin alpha |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)