













| General Information: |
|
| Name(s) found: |
UGA2 /
YBR006W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 497 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
response to oxidative stress
[IEP gamma-aminobutyric acid catabolic process [IEP glutamate decarboxylation to succinate [IGI |
| Molecular Function: |
succinate-semialdehyde dehydrogenase [NAD(P)+] activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTLSKYSKPT LNDPNLFRES GYIDGKWVKG TDEVFEVVDP ASGEIIARVP EQPVSVVEEA 60
61 IDVAYETFKT YKNTTPRERA KWLRNMYNLM LENLDDLATI ITLENGKALG EAKGEIKYAA 120
121 SYFEWYAEEA PRLYGATIQP LNPHNRVFTI RQPVGVCGII CPWNFPSAMI TRKAAAALAV 180
181 GCTVVIKPDS QTPLSALAMA YLAEKAGFPK GSFNVILSHA NTPKLGKTLC ESPKVKKVTF 240
241 TGSTNVGKIL MKQSSSTLKK LSFELGGNAP FIVFEDADLD QALEQAMACK FRGLGQTCVC 300
301 ANRLYVHSSI IDKFAKLLAE RVKKFVIGHG LDPKTTHGCV INSSAIEKVE RHKQDAIDKG 360
361 AKVVLEGGRL TELGPNFYAP VILSHVPSTA IVSKEETFGP LCPIFSFDTM EEVVGYANDT 420
421 EFGLAAYVFS KNVNTLYTVS EALETGMVSC NTGVFSDCSI PFGGVKESGF GREGSLYGIE 480
481 DYTVLKTITI GNLPNSI |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
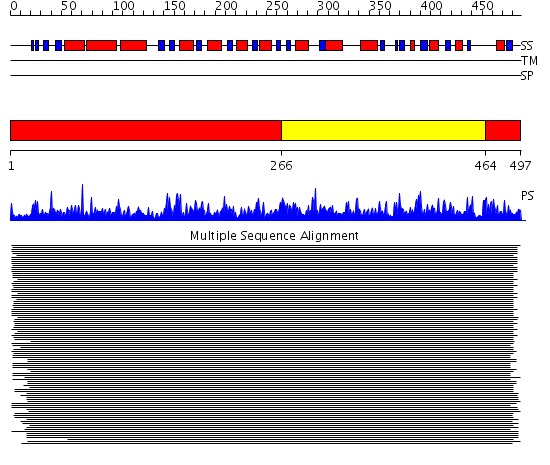
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..265] [464..497] |
1805.228787 | Aldehyde reductase (dehydrogenase), ALDH | |
| 2 | View Details | [266..463] | 1805.228787 | Aldehyde reductase (dehydrogenase), ALDH |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)