













| General Information: |
|
| Name(s) found: |
NOP9_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 655 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPKKRKTRGK KHSAKQKEEE VNSVVSPGIA KNDEGAGNDE GAYQVNRYTN EPVQPFFGAL 60
61 DPEEEKYFQQ AEQAFANHFT RENEDTRFFV DSIYREVQGK ELIVCNNVFG SKVLEKLFPL 120
121 STSRQIKSFF SALNGHYVEL IQTTFGSYAF EKLLSHMAAI LNLETQGVTE DQDDVLEGEN 180
181 ENVFMTAETL LMYMYNEIRP EVSMLMSHKL AVHVLGKIFL LLDGYRFVYH DGNRTTMESI 240
241 SVPESFPQFA YSIIDSATDN LDTPELRAYC VDKYASQIMR AFIRIDFERS KKTKKKHDPR 300
301 LVSKLLLSNE YDLKELPFME TLLKDETGSR ILEVIVENMS ASHLLRFYAV FEGRFYRLCV 360
361 HPIANFIMQR YIRRLGRKEI GSVIDELKKN TDNIIRKSFL PVLRTLLEKS NHLHCYQDDI 420
421 FSMIQTSVTN SGKDANIIPC LLRSKHKRDK ANPENKERKL VNNLLGAQLL EEMLHCEKQH 480
481 IQLLLDSVLD LSKEQILEYC LETVSSHLIE GILDLSDLNP VFLKKFLNIL SGSFATLAVS 540
541 APGSHIVDKA WKATRALPLY RTRIVRELAE GGDDVKFDFY GKKVCSNWKV ELFRRAPDEW 600
601 YRFMKQDEPS KRVHERARQY GRIVSGSSAN TQPLGEKRST ENDEELESGP KRVKV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
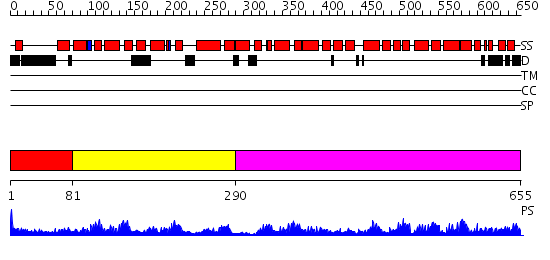
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [81..289] | 3.01 | Pumilio 1 | |
| 3 | View Details | [290..655] | 82.39794 | Pumilio 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)