













| General Information: |
|
| Name(s) found: |
GYRB_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 804 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNSYDSSSI KVLKGLDAVR KRPGMYIGDT DDGTGLHHMV FEVVDNAIDE ALAGHCKEII 60
61 VTIHADNSVS VQDDGRGIPT GIHPEEGVSA AEVIMTVLHA GGKFDDNSYK VSGGLHGVGV 120
121 SVVNALSQKL ELVIQREGKI HRQIYEHGVP QAPLAVTGET EKTGTMVRFW PSLETFTNVT 180
181 EFEYEILAKR LRELSFLNSG VSIRLRDKRD GKEDHFHYEG GIKAFVEYLN KNKTPIHPNI 240
241 FYFSTEKDGI GVEVALQWND GFQENIYCFT NNIPQRDGGT HLAGFRAAMT RTLNAYMDKE 300
301 GYSKKAKVSA TGDDAREGLI AVVSVKVPDP KFSSQTKDKL VSSEVKSAVE QQMNELLAEY 360
361 LLENPTDAKI VVGKIIDAAR AREAARRARE MTRRKGALDL AGLPGKLADC QERDPALSEL 420
421 YLVEGDSAGG SAKQGRNRKN QAILPLKGKI LNVEKARFDK MLSSQEVATL ITALGCGIGR 480
481 DEYNPDKLRY HSIIIMTDAD VDGSHIRTLL LTFFYRQMPE IVERGHVYIA QPPLYKVKKG 540
541 KQEQYIKDDE AMDQYQISIA LDGATLHTNA SAPALAGEAL EKLVSEYNAT QKMINRMERR 600
601 YPKAMLKELI YQPTLTEADL SDEQTVTRWV NALVSELNDK EQHGSQWKFD VHTNAEQNLF 660
661 EPIVRVRTHG VDTDYPLDHE FITGGEYRRI CTLGEKLRGL LEEDAFIERG ERRQPVASFE 720
721 QALDWLVKES RRGLSIQRYK GLGEMNPEQL WETTMDPESR RMLRVTVKDA IAADQLFTTL 780
781 MGDAVEPRRA FIEENALKAA NIDI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
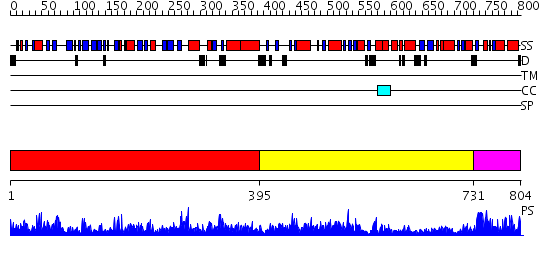
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..394] | 166.0 | DNA gyrase B | |
| 2 | View Details | [395..730] | 66.30103 | TOPOISOMERASE RESIDUES 410-1202, | |
| 3 | View Details | [731..804] | 1.62 | TOPOISOMERASE RESIDUES 410-1202, |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)