













| General Information: |
|
| Name(s) found: |
ODP2_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 630 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAIEIKVPDI GADEVEITEI LVKVGDKVEA EQSLITVEGD KASMEVPSPQ AGIVKEIKVS 60
61 VGDKTQTGAL IMIFDSADGA ADAAPAQAEE KKEAAPAAAP AAAAAKDVNV PDIGSDEVEV 120
121 TEILVKVGDK VEAEQSLITV EGDKASMEVP APFAGTVKEI KVNVGDKVST GSLIMVFEVA 180
181 GEAGAAAPAA KQEAAPAAAP APAAGVKEVN VPDIGGDEVE VTEVMVKVGD KVAAEQSLIT 240
241 VEGDKASMEV PAPFAGVVKE LKVNVGDKVK TGSLIMIFEV EGAAPAAAPA KQEAAAPAPA 300
301 AKAEAPAAAP AAKAEGKSEF AENDAYVHAT PLIRRLAREF GVNLAKVKGT GRKGRILRED 360
361 VQAYVKEAIK RAEAAPAATG GGIPGMLPWP KVDFSKFGEI EEVELGRIQK ISGANLSRNW 420
421 VMIPHVTHFD KTDITELEAF RKQQNEEAAK RKLDVKITPV VFIMKAVAAA LEQMPRFNSS 480
481 LSEDGQRLTL KKYINIGVAV DTPNGLVVPV FKDVNKKGII ELSRELMTIS KKARDGKLTA 540
541 GEMQGGCFTI SSIGGLGTTH FAPIVNAPEV AILGVSKSAM EPVWNGKEFV PRLMLPISLS 600
601 FDHRVIDGAD GARFITIINN TLSDIRRLVM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
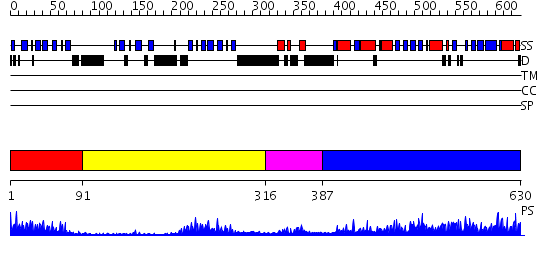
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..90] | 14.0 | Ipoyl domain of dihydrolipoamide acetyltransferase | |
| 2 | View Details | [91..315] | 5.4 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | |
| 3 | View Details | [316..386] | 24.69897 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | |
| 4 | View Details | [387..630] | 74.69897 | CRYSTALLOGRAPHIC AND ENZYMATIC INVESTIGATIONS ON THE ROLE OF SER558, HIS610 AND ASN614 IN THE CATALYTIC MECHANISM OF AZOTOBACTER VINELANDII DIHYDROLIPOAMIDE ACETYLTRANSFERASE (E2P) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)