













| General Information: |
|
| Name(s) found: |
SYA_SALTI
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Salmonella enterica subsp. enterica serovar Typhi |
| Length: | 876 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKSTAEIRQ AFLDFFHSKG HQVVASSSLV PNNDPTLLFT NAGMNQFKDV FLGLDKRNYS 60
61 RATTSQRCVR AGGKHNDLEN VGYTARHHTF FEMLGNFSFG DYFKHDAIQF AWELLTGENW 120
121 FALPKERLWV TVYETDDEAY EIWEKEVGIP RERIIRIGDN KGAPYASDNF WQMGDTGPCG 180
181 PCTEIFYDHG DHIWGGPPGS PEEDGDRYIE IWNIVFMQFN RQADGTMEPL PKPSVDTGMG 240
241 LERIAAVLQH VNSNYDIDLF RTLIEAVAKV TGSTDLGNKS LRVIADHIRS CAFLVADGVL 300
301 PSNENRGYVL RRIIRRAVRH GNMLGAKETF FYKLVGPLIE VMGSAGEELK RQQAQVEQVL 360
361 KTEEEQFART LERGLALLDE ELAKLQGDTL DGETAFRLYD TYGFPVDLTA DVCRERNIKV 420
421 DEAGFEAAME EQRRRAREAS GFGADYNAMI RVDSASEFKG YDHLELNGKV TALFVDGKAV 480
481 EAINAGQEAV VVLDQTPFYA ESGGQVGDKG ELKGAGFTFA VDDTQKYGQA IGHLGKLSAG 540
541 ALKVGDAVQA DVDEARRARI RLNHSATHLM HAALRQVLGT HVAQKGSLVS DKVLRFDFSH 600
601 NEAMKPSEIR EVEDLVNAQI RRNLPIETNI MDLDAAKAKG AMALFGEKYD ERVRVLSMGD 660
661 FSTELCGGTH ASRTGDIGLF RIISESGTAA GIRRIEAVTG EGAMATVHAQ SDRLNDIAHL 720
721 LKGDSQNLGD KVRAVLERTR QLEKELQQLK DQAAAQESAN LSSKAVDLNG VKLLVSELAG 780
781 IEPKMLRTMV DDLKNQLGST VIVLATVVEG KVSLIAGVSK DVTDRVKAGE LIGMVAQQVG 840
841 GKGGGRPDMA QAGGTDAAAL PAALASVQGW VSAKLQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
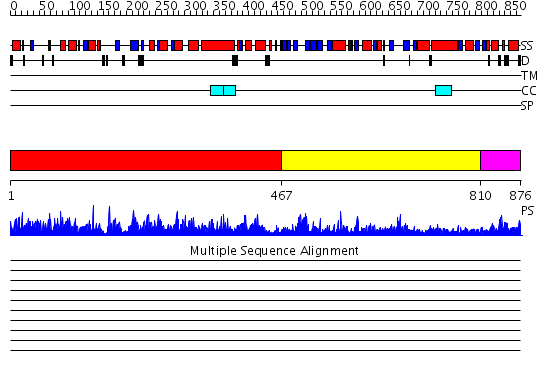
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..466] | 1000.0 | The crystal structure of the catalytic fragment of the alanyl-tRNA synthetase | |
| 2 | View Details | [467..809] | 58.0 | Structure of Staphylococcus aureus threonyl-tRNA synthetase complexed with an analogue of threonyl adenylate | |
| 3 | View Details | [810..876] | 12.853872 | No description for PF02272.10 was found. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)