













| General Information: |
|
| Name(s) found: |
gi|24374744
[NCBI NR]
gi|24349408 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 477 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
regulation of flagellum assembly
[ISS ciliary or flagellar motility [ISS |
| Molecular Function: |
transcription activator activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMQTDQRILL VGTPSERLSR LCCIFEFLGE QIDIITIEKL STCLQETRFR ALVISIDNMS 60
61 ADALKLVASQ YPWQPILLFG HVGELQVSNV LGHIEEPLNY PQLTELLHFC QVYGQVKRPQ 120
121 VPTSANQTKL FRSLVGRSDG IANVRHLISQ VATSDATVLV LGQSGTGKEV VARNIHYLSE 180
181 RRDGPFIPVN CGAIPPELLE SELFGHEKGS FTGAISSRKG RFELAEGGTL FLDEIGDMPL 240
241 QMQVKLLRVL QERVFERVGG TKTINADVRV VAATHRDLET MISSNEFRED LYYRLNVFPI 300
301 EMPALSERKD DVPLLLQELV SRVYNEGRGK VRFTQRAIES LKEHAWSGNV RELSNLVERL 360
361 TILYPGGLVD VNDLPIKYRH IDVPEYSVEL SEEQQERDAL ASIFTSEEPV EIPETRFPSE 420
421 LPPEGVNLKD LLAELEIDMI RQALEQQDNV VARAAEMLGI RRTTLVEKMR KYGMTKE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
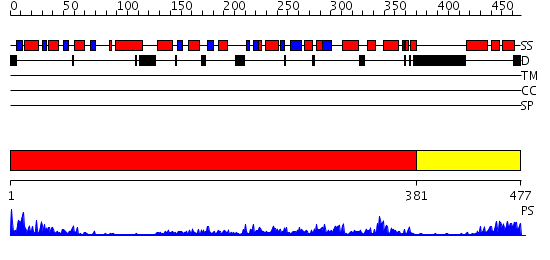
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..380] | 72.30103 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 2 | View Details | [381..477] | 69.154902 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)