













| General Information: |
|
| Name(s) found: |
gi|24372429
[NCBI NR]
gi|24346403 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 1517 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
lipid metabolic process
[ISS |
| Molecular Function: |
acetyl-CoA carboxylase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSNNQPKHM TQAQTPDPLF LQAEHLAKDF SLFPKHSKQS LSEEIKGLLN DDAIQANLKD 60
61 LASLDVDGYV AKVIQPTQDK GRPGAKRIIA DLKGRLITET HLGSFYSAEV ELNFGSRTRR 120
121 IGFIAQERTT ANGAWMPEHH YAACKAIRHF AELSMPIVYL IDTPGADAGE VANSQNQAHS 180
181 ISKAIAESAN VDVPTVGIVI GAGYSGGAIP LAAANILLSL RDGIFNTIQP QGLQSIARKY 240
241 NLSWQECAKS VGVSPEELYT SGCIDGIIDF SPGDRDERQH NLRRAIISSI EAVERAAVNF 300
301 VRESKDLREH YDRSLARFLN PSKNLMALEL NAELAVATSP TNHHNLFGSA YRYLRYLTLR 360
361 SRIHSISQEQ YGRLSRVSVP EGDLLARIQQ EQDRVFQAWL ASPDKLVYDE DLNKLWANFI 420
421 AKRDEISTER NMLTRFILGE PKENYKKARK ALLFNISWSL YHRWKSNAAN NFKGLIQHLE 480
481 NLPKSVTQAK WPELNQLTVL DVVVNDELRE DFIWQCHNIL IFNSLYDNVV GSLASIAKEA 540
541 MMSKSLSRNS VNNLLHKSID HALSTQDTAN DKAKFYKWLK YFMGQSNRAD LLVRVEQWKS 600
601 VGFPQLNDSL FVILTYFFER LLPEYFDSES DHSKYTGAIN PVRIGRRKDF WNRLTMGYQD 660
661 LLIQKVLRDE KRTGKMTWEN VIKQFFTHFD EINGDKMSAN LLNFPGFRLS IEDALDKGIR 720
721 PCGLITGIAD FKEKGQKVRV GLAVSNTAFQ AGAFDMASAE KFSALLIECA KRKLPVVCFI 780
781 SSGGMQTKEG AAALFSMAVV NDRITRFIRD NELPVLMFGF GDCTGGAQAS FVTHPLVQTY 840
841 YLSGTNMPFA GQMVVPAYLP STSTLSNYLS KVPGAMAGLV HNPFSENLDN QLASIDPLMP 900
901 MPTAKINEVI INALSTLVVE APAEEEEIVQ NDPRKLMKPI NKVLVHARGC TAVKLIRKAH 960
961 DNNINVVLVA SDPDMTAVPA DMLKESDKLV CLGGNTSDES YLNAYSVLKV AEYEQVDALH1020
1021 PGIGFLSESP QFAALCVNNG VNFVGPSVHS MTTMGNKSNA IKTSQAQNVP VVPGSHGILT1080
1081 NAEQAVNVAS EIGYPVLLKA VQGGGGKGIQ VVKRPEDMIG LFQKTATEAA AAFGNGDLYL1140
1141 EKYVTSLRHI EVQLLRDKFG HAKVLGLRDC SVQRNNQKVV EESGSTMLPD ELKKQVLAYT1200
1201 RALGDATDYM GAGTVEFIYN LDANEVYFME MNTRLQVEHP VTEATSGIDI VSAQFDIAAG1260
1261 RSIEHLEPKE IGYAMEVRVT AEKAALDSHG VLQLIPNPGK ITECVFPDHP DVEIISIAAP1320
1321 GKEVSPYYDS LIAQVIMRGE NREDVIAKLH AYLDSVVLKG IATNIPLLKL ILSDGTFKEG1380
1381 VYDTNYLPRF MAELDIPALI AEMEAAAETV GVDTESLRVG ESNELKVLAQ GAGIFYTSPA1440
1441 PGEPDFVKEG DIVTAEQTLA LTEAMKMFSQ VNLAGFNRQG AVLYPEDKKY RIERILNSNG1500
1501 QQVSQGDLLF VVSPVED |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
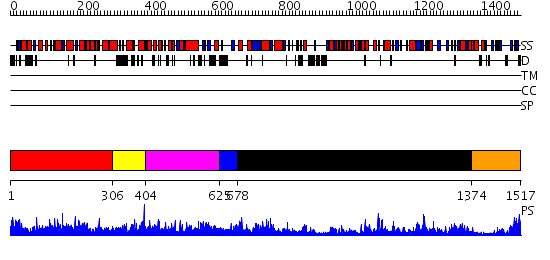
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..305] | 85.39794 | Crystal structure of propionyl-CoA carboxylase, beta subunit (TM0716) from THERMOTOGA MARITIMA at 2.30 A resolution | |
| 2 | View Details | [306..403] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [404..624] | 1.019963 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [625..677] | 53.69897 | No description for 2f9yB was found. | |
| 5 | View Details | [678..1373] | 90.39794 | Carbamoyl phosphate synthetase, large subunit connection domain; Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain; Carbamoyl phosphate synthetase (CPS), large subunit PreATP-grasp domains; Carbamoyl phosphate synthetase (CPS), large subunit ATP-binding domains | |
| 6 | View Details | [1374..1517] | 119.0 | No description for 2qf7A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)