













| General Information: |
|
| Name(s) found: |
VPS3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1011 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKKKTNNDK GKEVKENEGK LDIDSESSPH ERENDKKKTE DDSLRATESE ETNTHNANPN 60
61 ETVRADKFSQ EESRPIEDSP HTDKNTAQES CQPSSAEDNV INTDITSLNE KTSTNDEQEK 120
121 GLPLKISEGP FTISTLLDNV PSDLIYTCCE AYENHIFLGT TTGDLLHYFE LERGNYMLVS 180
181 QTKFDAESNS KIDKILLLPK VEGALILCDN ELVLFILPEF APRPNTTRLK GISDVVICNF 240
241 SRSSKAYRIY AFHAEGVRLL KISADSLVLT KAFNFKLIDK ACAHEETLMV SKLNSYELIN 300
301 LKSSQVIPLF RISETDEDLE PIITSFNEQS EFLVCSGGGS YDSGAMALVV NHHGDIIKGT 360
361 IVLKNYPRNV IVEFPYIIAE SAFQSVDIYS ALPSEKSQLL QSITTSGSDL KISKSDNVFT 420
421 NTNNSEEFKE KIFNKLRLEP LTHSDNKFRI ERERAFVEES YEEKTSLIVY NNLGIHLLVP 480
481 TPMVLRFTSC EESEIDNIED QLKKLAKKDL TKFEHIEAKY LMSLLLFLMT LHYDHIEDEV 540
541 MKKWCDFSDK VDIRILFYMF GWKVYSEIWC FHGLINIVER LKSLKLTNKC ENILKMLLMM 600
601 KNELKKKNKT GLLTNDFDDI MKTIDITLFK LRLEKKETIT VDMFERESYD EIIREINLHD 660
661 DKLPRIELLI EIYKEKGEYL KALNLLREAG DYISLVSFIE ENLKKLPEDY IKERIADDLL 720
721 LTLKQGDENT EECAIKKVLK ILDMACINKN DFLNKIPAEE TSLKVSFIEQ LGVQNSNDSK 780
781 FLFNYYLAKL REIINQSNIW SILGDFIKEY KDDFAYDKTD ITNFIHIKLK HSLQCENFSK 840
841 YYEKCENLKS ENEKDDEFIN FTFDEISKID KEHILTLLFF PNELTNWVSS EELLKIYLSF 900
901 NDFRSVEKYI GKQNLVAVMK QYLDISSLNY SVELVTNLLQ RNFELLDDTD IQLKILETIP 960
961 SVFPVQTISE LLLKVLIKYQ EKKEESNLRK CLLKNQISIS DELSRNFDSQ G |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
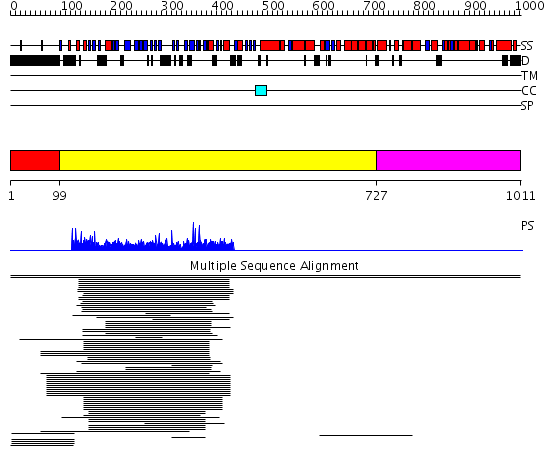
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [99..726] | 15.038994 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [727..1011] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [407..491] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [492..1011] | 0.962 | Clathrin heavy chain proximal leg segment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)