













| General Information: |
|
| Name(s) found: |
GDE1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1223 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKFGKTFANH RIPEWSSQYV GYKSLKKMIK EITRLQEDIY RAHNKNSYDE GRPPTKMRDS 60
61 SNSAQNYLDS PKIQKLLASF FFAVDRDIEK VDTFYNSQYA EYKKRFERLL SSNQFNEIKS 120
121 TLVVDANKED AVAQTLLTKD TREMNMLLKG TSQASRLSYH KDDLIEIQSI LAELRKQFRN 180
181 LKWYAELNKR AFGKILKKLD KKVGTNQQMS TMKTRILPLQ FANDSLITKD LSLLKTIWEQ 240
241 VTFRINSYER VMRSTSPNAN ANDNTEFFKI ICVFIEEDDS KGLIRELTNL YSELSLIPTR 300
301 IMISVLNKAA LSKSLACIDA ILKVIPSLND SEDINRRNFF HHHIIAIGKL IRKQEILSRK 360
361 KKSQPSKYTN SEGEIVTDLR TLHTTLSAPA ESDSITEEEK SSACTLSYIL EELPIHLRPC 420
421 LFQHDNYKRT PLHYSCQYGL SEVTKLIIKL MKEWNIWNEI PIDDVSAFGD AESLTPLHLC 480
481 VLGAHPKTTE VLLQSLDPNV KLKSSSLLHL ATEWNNYPLL HVLLSSKRFD INYQDNELHE 540
541 TPLYLACRLN FFEAAVCLLY NGADLEIREK LFGWTAIFVA AAEGFTDIVK LLIANNANFD 600
601 IEDEGGWTPM EHAVLRGHLH IADMVQIRDE LVTHPHSQLN SGSEEKEPLN EISAGELNER 660
661 NENGNGGNKG SLGKLAGPIK SYGHRFLDNN ESLILITLGS NDTRNKSPSI SLSSEALAKV 720
721 IGLETDCALS LVISCNDSID KSSVILDLPL DDNVDAVDFK VPFKVDYSHT LYFDIVPTYG 780
781 TRSLETHNRI DCQKNNNNYV MARGVSMLNK SYSSVGVNRS ILNGSVTVPI IANHTLEILG 840
841 TLKFEYIIIT PFEHPQLPLE RTETYWKSLV STRVIGHRGL GKNNPNKSLQ IGENTVESFI 900
901 MAASLGASYV EFDVQLTKDN VPVVYHDFLV AETGVDIPMH ELTLEQFLDL NNADKEHIQR 960
961 GAGHSPHHVN GADTALQKYR GRSVDDSDVS TLRRAWDLHD NDPNGKSNNA HWSDNRMRLT1020
1021 KTFKKNNFKG NARGHSIASS FVTLKELFKK IPANVGFNIE CKFPMLDEAE EEELGQIMME1080
1081 MNHWVDTVLK VVFDNANGRD IIFSSFHPDI CIMLSLKQPV IPILFLTEGG SEQMADLRAS1140
1141 SLQNGIRFAK KWNLLGIVSA AAPILKAPRL VQVVKSNGLV CVTYGVDNND PENASIQIEA1200
1201 GVDAVIVDSV LAIRRGLTKK NEK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
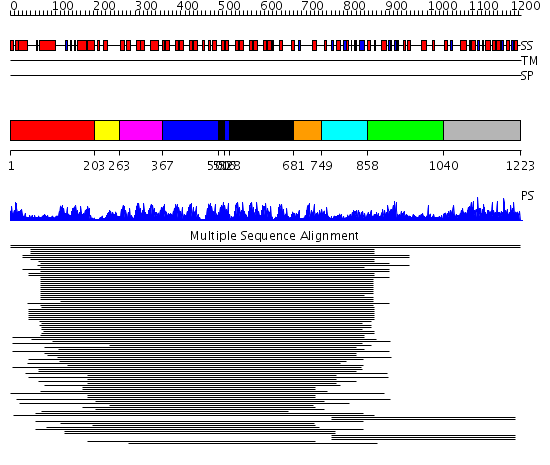
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..202] | 50.102373 | SPX domain No confident structure predictions are available. | |
| 2 | View Details | [203..262] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [263..366] | 177.045757 | Ankyrin-R | |
| 4 | View Details | [367..499] [516..527] |
177.045757 | Ankyrin-R | |
| 5 | View Details | [500..515] [528..680] |
177.045757 | Ankyrin-R | |
| 6 | View Details | [681..748] | 9.0 | Swi6 ankyrin-repeat fragment | |
| 7 | View Details | [749..857] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [858..1039] | 47.69897 | Hypothetical protein TM1621 | |
| 9 | View Details | [1040..1223] | 14.130768 | Glycerophosphoryl diester phosphodiesterase family No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)