













| General Information: |
|
| Name(s) found: |
gi|47097258
[NCBI NR]
gi|47014358 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Listeria monocytogenes serotype 1/2a str. F6854 |
| Length: | 648 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDDTEKRVHK YIEKHDLIRS DDKLLVAVSG GPDSLALLHF LWNSNLVPKE AISVAHLNHQ 60
61 LRENAAKEQR VVETFCERQG IPFYIEEVDI KSRAQSLQKG LEETARIVRY DFFEKVMAEK 120
121 NINKLVLAHH ADDQIETILM RLVRGSASIG WSGIQPKREL KGGQAIRPFL PITKAEIIDY 180
181 AQKHELAYEI DESNTSQEYT RNRYRAQLLP FLKQENPAVY SHFERFSEET SEDFQFLEAL 240
241 ASDLLKKNLI KNGKQTTLLL SSFKNEANPL QRRAIHLLLR YLYNEDASFI TVNHIYQIIQ 300
301 MIQSDNPSSS IDLPNKLIAN RAYDKLHFQF GEREAPSEFY HQLELNDRIE LDNKASIRLK 360
361 LKSSVVQTNG LNGMLLDAEE ITLPLIVRNR VNGDRMTMKG QAGSKKLKDI FIDAKIPRQE 420
421 RDKLPVITDY TGKILWVPGV KKSAYDREFS RSKKQYIIRY TRNIGGNESM HNDIQKVLIS 480
481 EDELQEKIRE LGRELTTEYE GRNPLVVGVL KGATPFMTDL LKRVDTYLEM DFMDVSSYGN 540
541 GTVSSGEVKI IKDLNASVEG RDVLVIEDII DSGRTLSYLV DLIKYRKAKS VKLVTLLDKP 600
601 AGRNVEIEAD YVGFVVPNEF VVGYGLDYAE RYRNLPYIGI LKPEIYSE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
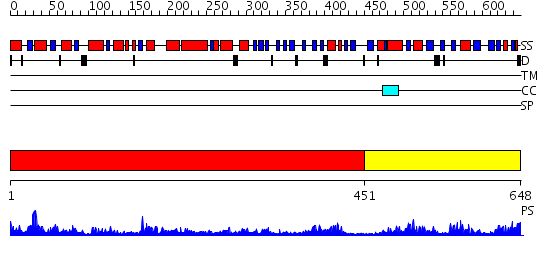
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..450] | 59.30103 | Putative cell cycle protein MesJ, N-terminal domain; Putative cell cycle protein MesJ, middle and C-terminal domain | |
| 2 | View Details | [451..648] | 35.045757 | Crystal Structure of Hypoxanthine-Guanine Phosphoribosyltransferase from Thermoanaerobacter tengcongensis |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)